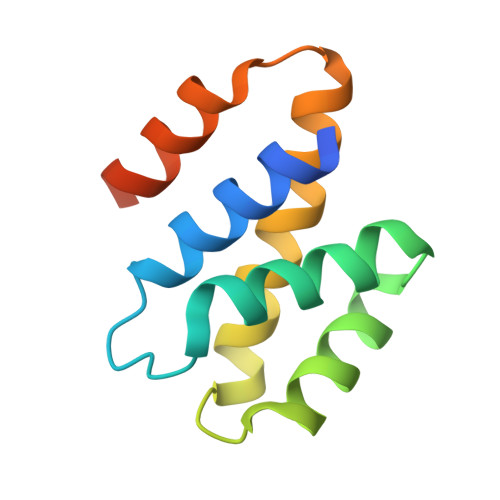
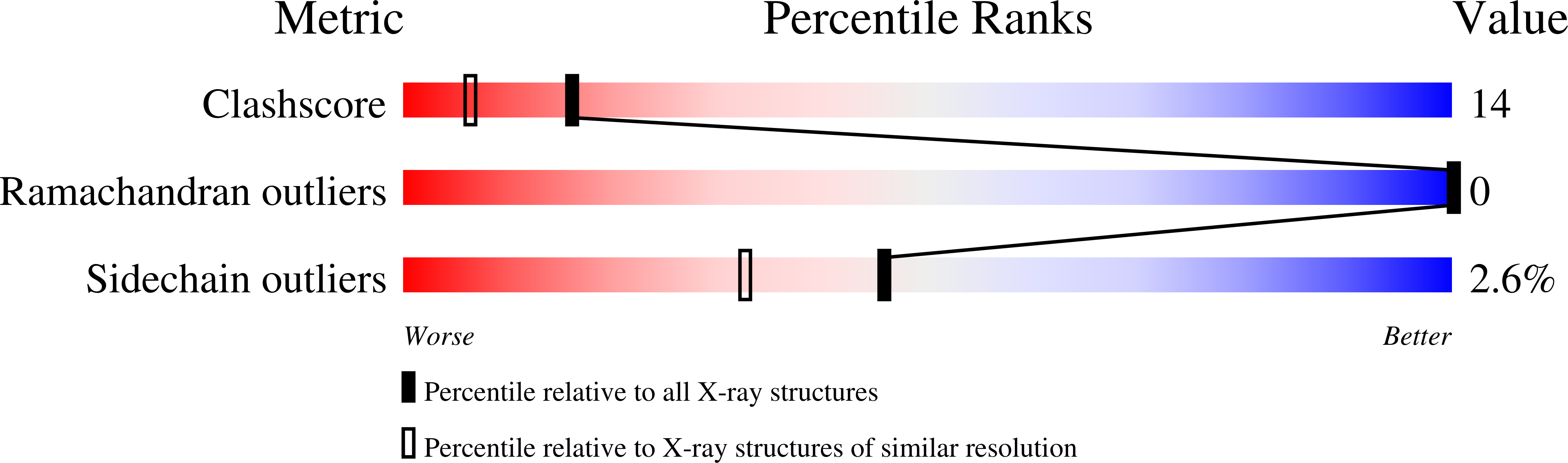Structure of the N-Terminal Mlp1-Binding Domain of the Saccharomyces Cerevisiae Mrna-Binding Protein, Nab2.
Grant, R.P., Marshall, N.J., Yang, J.-C., Fasken, M.B., Kelly, S.M., Harreman, M.T., Neuhaus, D., Corbett, A.H., Stewart, M.(2008) J Mol Biol 376: 1048
- PubMed: 18190927
- DOI: https://doi.org/10.1016/j.jmb.2007.11.087
- Primary Citation of Related Structures:
2JPS, 2V75 - PubMed Abstract:
Nuclear abundant poly(A) RNA-binding protein 2 (Nab2) is an essential yeast heterogeneous nuclear ribonucleoprotein that modulates both mRNA nuclear export and poly(A) tail length. The N-terminal domain of Nab2 (residues 1-97) mediates interactions with both the C-terminal globular domain of the nuclear pore-associated protein, myosin-like protein 1 (Mlp1), and the mRNA export factor, Gfd1. The solution and crystal structures of the Nab2 N-terminal domain show a primarily helical fold that is analogous to the PWI fold found in several other RNA-binding proteins. In contrast to other PWI-containing proteins, we find no evidence that the Nab2 N-terminal domain binds to nucleic acids. Instead, this domain appears to mediate protein:protein interactions that facilitate the nuclear export of mRNA. The Nab2 N-terminal domain has a distinctive hydrophobic patch centered on Phe73, consistent with this region of the surface being a protein:protein interaction site. Engineered mutations within this hydrophobic patch attenuate the interaction with the Mlp1 C-terminal domain but do not alter the interaction with Gfd1, indicating that this patch forms a crucial component of the interface between Nab2 and Mlp1.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH, UK.