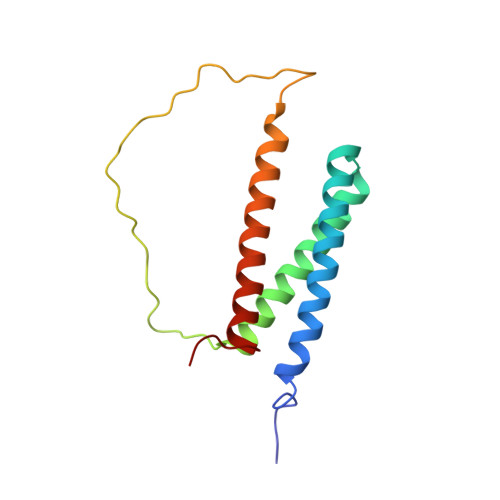
Structural interaction between DISC1 and ATF4 underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders.
Wang, X., Ye, F., Wen, Z., Guo, Z., Yu, C., Huang, W.K., Rojas Ringeling, F., Su, Y., Zheng, W., Zhou, G., Christian, K.M., Song, H., Zhang, M., Ming, G.L.(2019) Mol Psychiatry
- PubMed: 31444471
- DOI: https://doi.org/10.1038/s41380-019-0485-2
- Primary Citation of Related Structures:
6IRR - PubMed Abstract:
Psychiatric disorders are a collection of heterogeneous mental disorders arising from a contribution of genetic and environmental insults, many of which molecularly converge on transcriptional dysregulation, resulting in altered synaptic functions. The underlying mechanisms linking the genetic lesion and functional phenotypes remain largely unknown. Patient iPSC-derived neurons with a rare frameshift DISC1 (Disrupted-in-schizophrenia 1) mutation have previously been shown to exhibit aberrant gene expression and deficits in synaptic functions. How DISC1 regulates gene expression is largely unknown. Here we show that Activating Transcription Factor 4 (ATF4), a DISC1 binding partner, is more abundant in the nucleus of DISC1 mutant human neurons and exhibits enhanced binding to a collection of dysregulated genes. Functionally, overexpressing ATF4 in control neurons recapitulates deficits seen in DISC1 mutant neurons, whereas transcriptional and synaptic deficits are rescued in DISC1 mutant neurons with CRISPR-mediated heterozygous ATF4 knockout. By solving the high-resolution atomic structure of the DISC1-ATF4 complex, we show that mechanistically, the mutation of DISC1 disrupts normal DISC1-ATF4 interaction, and results in excessive ATF4 binding to DNA targets and deregulated gene expression. Together, our study identifies the molecular and structural basis of an DISC1-ATF4 interaction underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders.
Organizational Affiliation:
School of Basic Medical Sciences, Fudan University, 200032, Shanghai, China.