Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Bastard, K., Isabet, T., Stura, E.A., Legrand, P., Zaparucha, A.(2018) Sci Rep 8: 16587-16587
- PubMed: 30410048
- DOI: https://doi.org/10.1038/s41598-018-34795-9
- Primary Citation of Related Structures:
6EUO, 6EUR, 6EXF, 6EXH, 6F2A, 6F2B, 6F2E, 6F6J, 6F9P - PubMed Abstract:
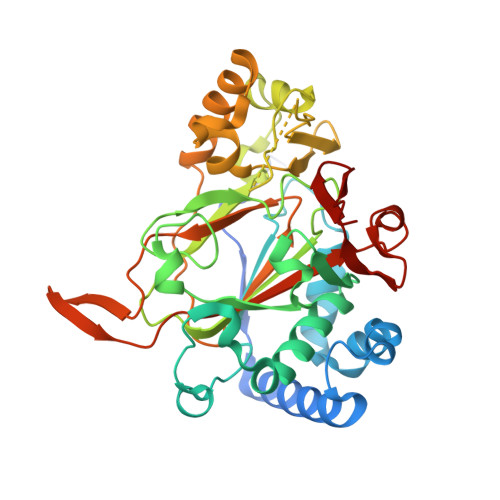
Iron(II)/α-ketoacid-dependent oxygenases (αKAOs) are enzymes that catalyze the oxidation of unactivated C-H bonds, mainly through hydroxylation. Among these, those that are active towards amino-acids and their derivatives are grouped in the Clavaminate Synthase Like (CSL) family. CSL enzymes exhibit high regio- and stereoselectivities with strict substrate specificity. This study reports the structural elucidation of two new regiodivergent members, KDO1 and KDO5, active towards lysine, and the structural and computational analysis of the whole family through modelling and classification of active sites. The structures of KDO1 and KDO5 in complex with their ligands show that one exact position in the active site controls the regioselectivity of the reaction. Our results suggest that the substrate specificity and high stereoselectivity typical of this family is linked to a lid that closes up in order to form a sub-pocket around the side chain of the substrate. This dynamic lid is found throughout the family with varying sequence and length and is associated with a conserved stable dimeric interface. Results from this study could be a starting-point for exploring the functional diversity of the CSL family and direct in vitro screening in the search for new enzymatic activities.
Organizational Affiliation:
Génomique Métabolique, Genoscope, Institut François Jacob, CEA, CNRS, Univ Evry, Université Paris-Saclay, 91057, Evry, France.