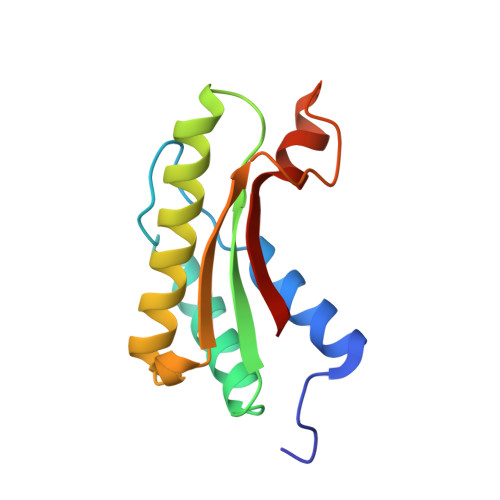
Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability.
Dai, S., Sun, C., Tan, K., Ye, S., Zhang, R.(2017) Cell Calcium 66: 78-89
- PubMed: 28807152
- DOI: https://doi.org/10.1016/j.ceca.2017.05.016
- Primary Citation of Related Structures:
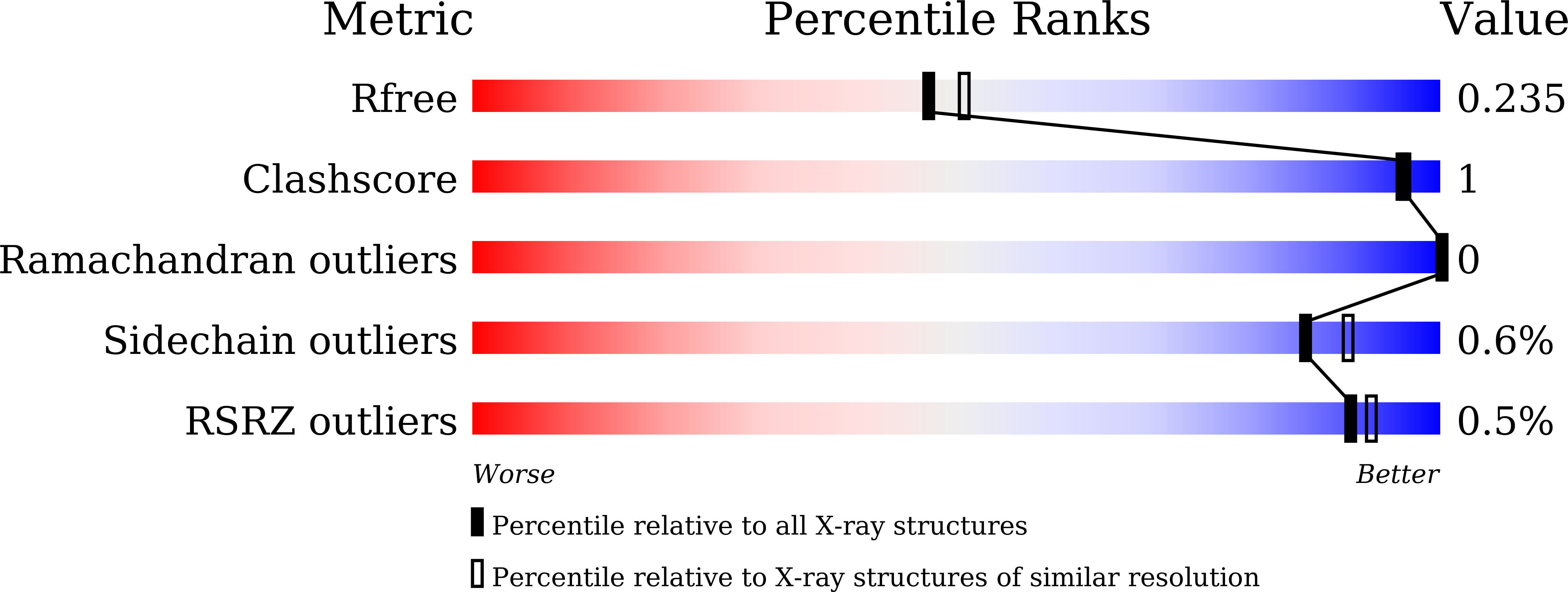
5WTL, 5WTP - PubMed Abstract:
Eukaryotic thrombospondin type 3 repeat (TT3R) is an efficient calcium ion (Ca 2+ ) binding motif only found in mammalian thrombospondin family. TT3R has also been found in prokaryotic cellulase Cel5G, which was thought to forfeit the Ca 2+ -binding capability due to the formation of intra-repeat disulfide bonds, instead of the inter-repeat ones possessed by eukaryotic TT3Rs. In this study, we have identified an enormous number of prokaryotic TT3R-containing proteins belonging to several different protein families, including outer membrane protein A (OmpA), an important structural protein connecting the outer membrane and the periplasmic peptidoglycan layer in gram-negative bacteria. Here, we report the crystal structure of the periplasmic region of OmpA from Capnocytophaga gingivalis, which contains a linker region comprising five consecutive TT3Rs. The structure of OmpA-TT3R exhibits a well-ordered architecture organized around two tightly-coordinated Ca 2+ and confirms the presence of abnormal intra-repeat disulfide bonds. Further mutagenesis studies showed that the Ca 2+ -binding capability of OmpA-TT3R is indeed dependent on the proper formation of intra-repeat disulfide bonds, which help to fix a conserved glycine residue at its proper position for Ca 2+ coordination. Additionally, despite lacking inter-repeat disulfide bonds, the interfaces between adjacent OmpA-TT3Rs are enhanced by both hydrophobic and conserved aromatic-proline interactions.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.