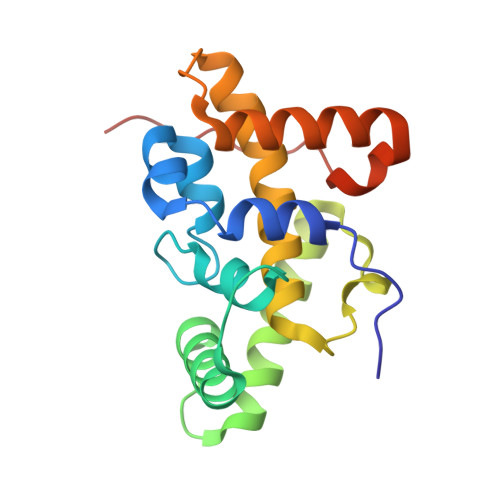
Rift Valley fever phlebovirus NSs protein core domain structure suggests molecular basis for nuclear filaments.
Barski, M., Brennan, B., Miller, O.K., Potter, J.A., Vijayakrishnan, S., Bhella, D., Naismith, J.H., Elliott, R.M., Schwarz-Linek, U.(2017) Elife 6
- PubMed: 28915104
- DOI: https://doi.org/10.7554/eLife.29236
- Primary Citation of Related Structures:
5OOO - PubMed Abstract:
Rift Valley fever phlebovirus (RVFV) is a clinically and economically important pathogen increasingly likely to cause widespread epidemics. RVFV virulence depends on the interferon antagonist non-structural protein (NSs), which remains poorly characterized. We identified a stable core domain of RVFV NSs (residues 83-248), and solved its crystal structure, a novel all-helical fold organized into highly ordered fibrils. A hallmark of RVFV pathology is NSs filament formation in infected cell nuclei. Recombinant virus encoding the NSs core domain induced intranuclear filaments, suggesting it contains all essential determinants for nuclear translocation and filament formation. Mutations of key crystal fibril interface residues in viruses encoding full-length NSs completely abrogated intranuclear filament formation in infected cells. We propose the fibrillar arrangement of the NSs core domain in crystals reveals the molecular basis of assembly of this key virulence factor in cell nuclei. Our findings have important implications for fundamental understanding of RVFV virulence.
Organizational Affiliation:
Biomedical Sciences Research Complex, University of St Andrews, St Andrews, United Kingdom.