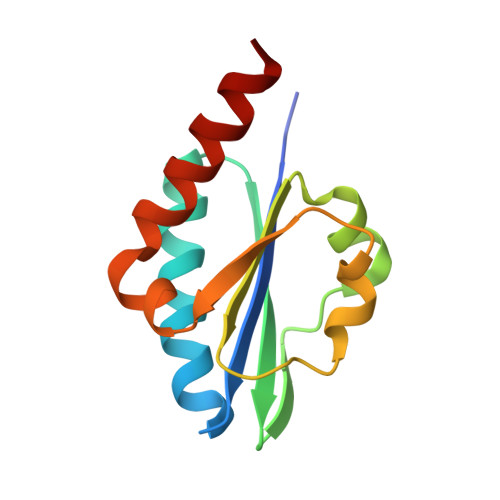
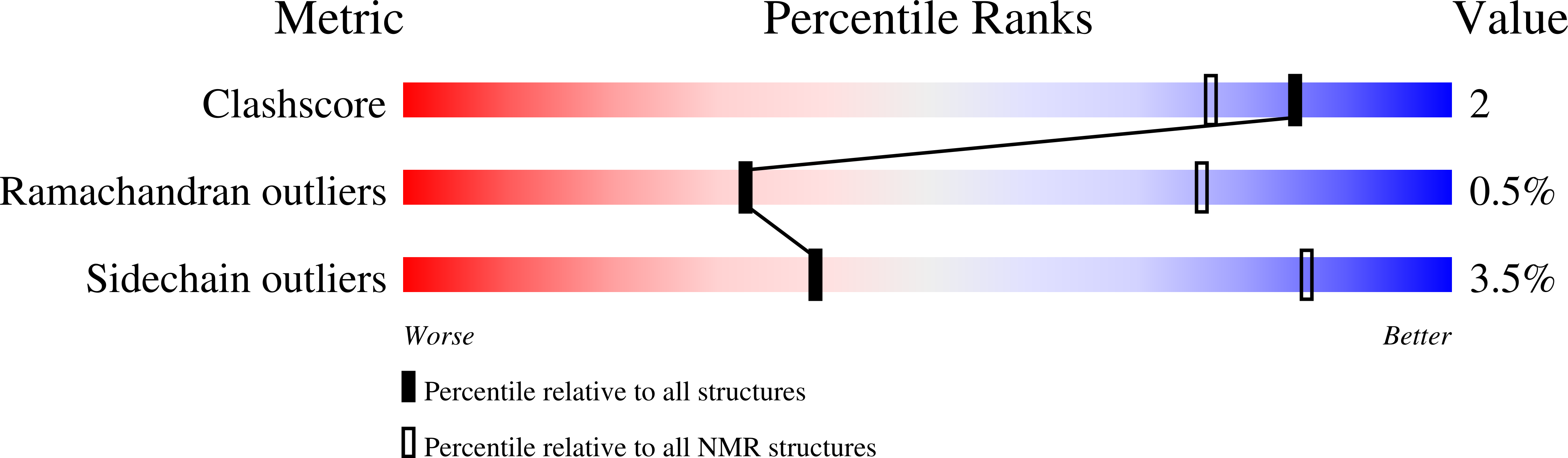Solution structure of Enzyme IIB subunit of PTS system from Escherichia coli K12, Northeast Structural Genomics Consortium target ER315/Ontario Center for Structural Proteomics target ec0544
Wu, B., Yee, A., Gutmanas, A., Semest, A., Arrowsmith, C.H.To be published.