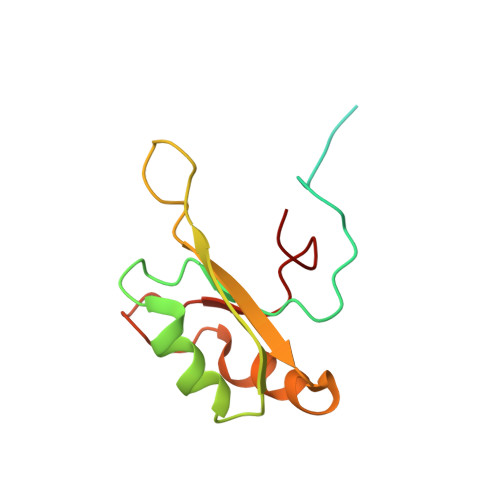
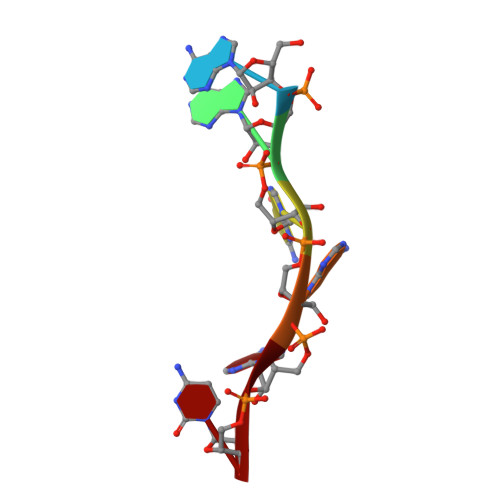
Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1
Clery, A., Jayne, S., Benderska, N., Dominguez, C., Stamm, S., Allain, F.H.(2011) Nat Struct Mol Biol 18: 443-450
- PubMed: 21399644
- DOI: https://doi.org/10.1038/nsmb.2001
- Primary Citation of Related Structures:
2KXN - PubMed Abstract:
Tra2-β1 is a unique splicing factor as its single RNA recognition motif (RRM) is located between two RS (arginine-serine) domains. To understand how this protein recognizes its RNA target, we solved the structure of Tra2-β1 RRM in complex with RNA. The central 5'-AGAA-3' motif is specifically recognized by residues from the β-sheet of the RRM and by residues from both extremities flanking the RRM. The structure suggests that RNA binding by Tra2-β1 induces positioning of the two RS domains relative to one another. By testing the effect of Tra2-β1 and RNA mutations on the splicing of SMN2 exon 7, we validated the importance of the RNA-protein contacts observed in the structure for the function of Tra2-β1 and determined the functional sequence of Tra2-β1 in SMN2 exon 7. Finally, we propose a model for the assembly of multiple RNA binding proteins on this exon.
Organizational Affiliation:
Institute for Molecular Biology and Biophysics, Swiss Federal Institute of Technology, Zürich, Switzerland.