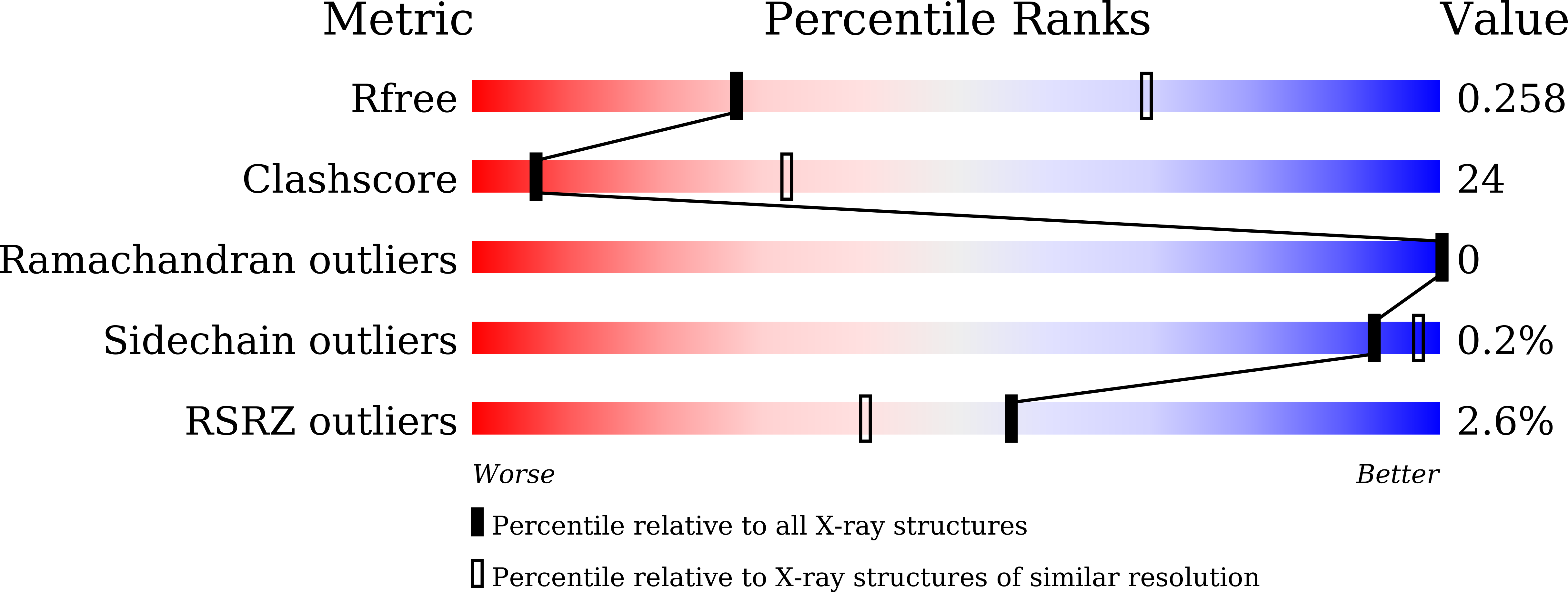
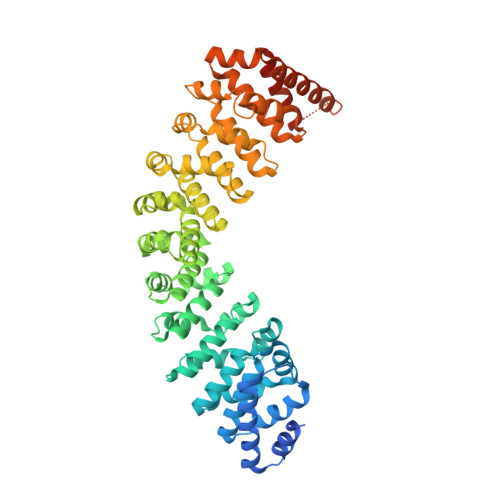
Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Park, J., Kim, H.I., Jeong, H., Lee, M., Jang, S.H., Yoon, S.Y., Kim, H., Park, Z.Y., Jun, Y., Lee, C.(2020) Autophagy 16: 991-1006
- PubMed: 31512555
- DOI: https://doi.org/10.1080/15548627.2019.1659615
- Primary Citation of Related Structures:
6KBM, 6KBN - PubMed Abstract:
Armadillo (ARM) repeat proteins constitute a large protein family with diverse and fundamental functions in all organisms, and armadillo repeat domains share high structural similarity. However, exactly how these structurally similar proteins can mediate diverse functions remains a long-standing question. Vac8 (vacuole related 8) is a multifunctional protein that plays pivotal roles in various autophagic pathways, including piecemeal microautophagy of the nucleus (PMN) and cytoplasm-to-vacuole targeting (Cvt) pathways in the budding yeast Saccharomyces cerevisiae . Vac8 comprises an H1 helix at the N terminus, followed by 12 armadillo repeats. Herein, we report the crystal structure of Vac8 bound to Atg13, a key component of autophagic machinery. The 70-Å extended loop of Atg13 binds to the ARM domain of Vac8 in an antiparallel manner. Structural, biochemical, and in vivo experiments demonstrated that the H1 helix of Vac8 intramolecularly associates with the first ARM and regulates its self-association, which is crucial for Cvt and PMN pathways. The structure of H1 helix-deleted Vac8 complexed with Atg13 reveals that Vac8[Δ19-33]-Atg13 forms a heterotetramer and adopts an extended superhelical structure exclusively employed in the Cvt pathway. Most importantly, comparison of Vac8-Nvj1 and Vac8-Atg13 provides a molecular understanding of how a single ARM domain protein adopts different quaternary structures depending on its associated proteins to differentially regulate 2 closely related but distinct cellular pathways.
Organizational Affiliation:
Department of Biological Sciences, School of Life Sciences, Ulsan National Institute of Science and Technology , Ulsan, Republic of Korea.