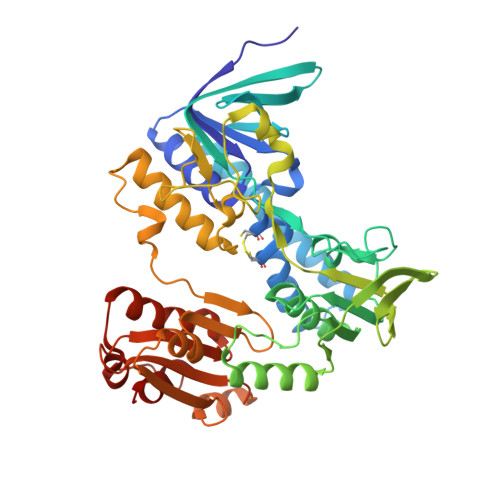
Hypothiocyanous acid reductase is critical for host colonization and infection by Streptococcus pneumoniae.
Shearer, H.L., Currie, M.J., Agnew, H.N., Trappetti, C., Stull, F., Pace, P.E., Paton, J.C., Dobson, R.C.J., Dickerhof, N.(2024) J Biol Chem : 107282-107282
- PubMed: 38604564
- DOI: https://doi.org/10.1016/j.jbc.2024.107282
- Primary Citation of Related Structures:
8UUB - PubMed Abstract:
The major human pathogen Streptococcus pneumoniae encounters the immune-derived oxidant hypothiocyanous acid (HOSCN) at sites of colonization and infection. We recently identified the pneumococcal hypothiocyanous acid reductase (Har), a member of the flavoprotein disulfide reductase enzyme family, and showed that it contributes to the HOSCN tolerance of S. pneumoniae in vitro. Here, we demonstrate in mouse models of pneumococcal infection that Har is critical for colonization and invasion. In a colonization model, bacterial load was attenuated dramatically in the nasopharynx when har was deleted in S. pneumoniae. The Δhar strain was also less virulent compared to wild type in an invasion model as reflected by a significant reduction in bacteria in the lungs and no dissemination to the blood and brain. Kinetic measurements with recombinant Har demonstrated that this enzyme reduced HOSCN with near diffusion limited catalytic efficiency, using either NADH (k cat /K M = 1.2 x 10 8 M -1 s -1 ) or NADPH (k cat /K M = 2.5 x 10 7 M -1 s -1 ) as electron donors. We determined the X-ray crystal structure of Har in complex with the FAD cofactor to 1.50 Å resolution, highlighting the active site architecture characteristic for this class of enzymes. Collectively, our results demonstrate that pneumococcal Har is a highly efficient HOSCN reductase, enabling survival against oxidative host immune defenses. In addition, we provide structural insights that may aid the design of Har inhibitors.
Organizational Affiliation:
Mātai Hāora - Centre for Redox Biology and Medicine, Department of Pathology and Biomedical Science, University of Otago Christchurch, Christchurch, New Zealand; Biomolecular Interaction Centre, MacDiarmid Institute for Advanced Materials and Nanotechnology and School of Biological Sciences, University of Canterbury, Christchurch, New Zealand; Maurice Wilkins Centre for Molecular Biodiscovery, New Zealand.