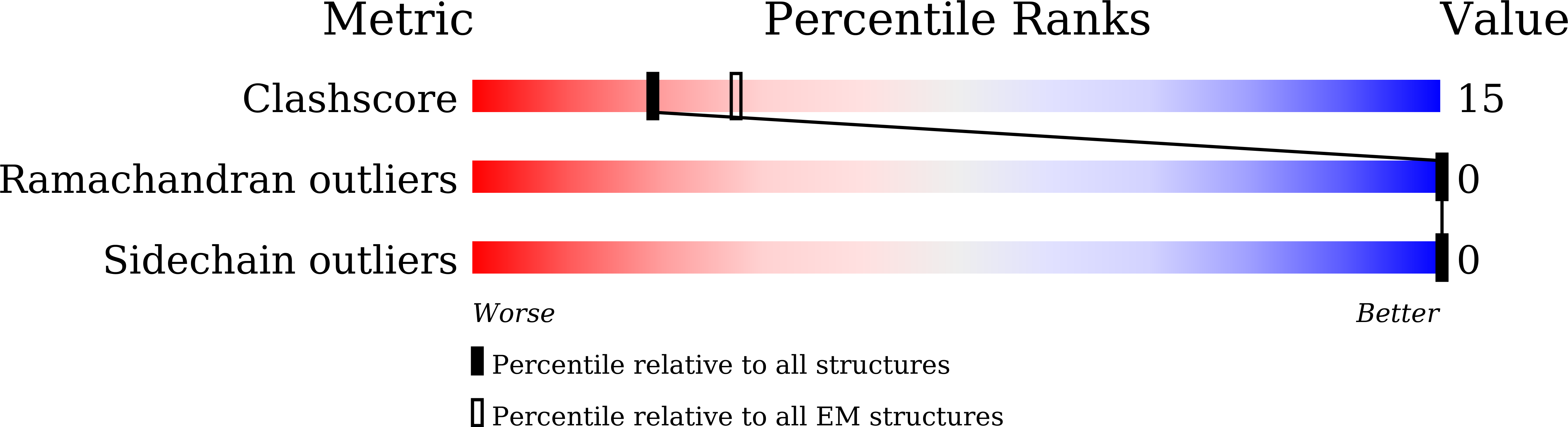Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Li, Z., Xia, H., Rao, G., Fu, Y., Chong, T., Tian, K., Yuan, Z., Cao, S.(2024) Nat Commun 15: 2284-2284
- PubMed: 38480794
- DOI: https://doi.org/10.1038/s41467-024-46624-x
- Primary Citation of Related Structures:
8K42, 8K43, 8K44, 8K49, 8K4A, 8W9P, 8W9Q, 8W9R - PubMed Abstract:
Banna virus (BAV) is the prototype Seadornavirus, a class of reoviruses for which there has been little structural study. Here, we report atomic cryo-EM structures of three states of BAV virions-surrounded by 120 spikes (full virions), 60 spikes (partial virions), or no spikes (cores). BAV cores are double-layered particles similar to the cores of other non-turreted reoviruses, except for an additional protein component in the outer capsid shell, VP10. VP10 was identified to be a cementing protein that plays a pivotal role in the assembly of BAV virions by directly interacting with VP2 (inner capsid), VP8 (outer capsid), and VP4 (spike). Viral spikes (VP4/VP9 heterohexamers) are situated on top of VP10 molecules in full or partial virions. Asymmetrical electrostatic interactions between VP10 monomers and VP4 trimers are disrupted by high pH treatment, which is thus a simple way to produce BAV cores. Low pH treatment of BAV virions removes only the flexible receptor binding protein VP9 and triggers significant conformational changes in the membrane penetration protein VP4. BAV virions adopt distinct spatial organization of their surface proteins compared with other well-studied reoviruses, suggesting that BAV may have a unique mechanism of penetration of cellular endomembranes.
Organizational Affiliation:
CAS Key Laboratory of Special Pathogens, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences, Wuhan, 430071, PR China.