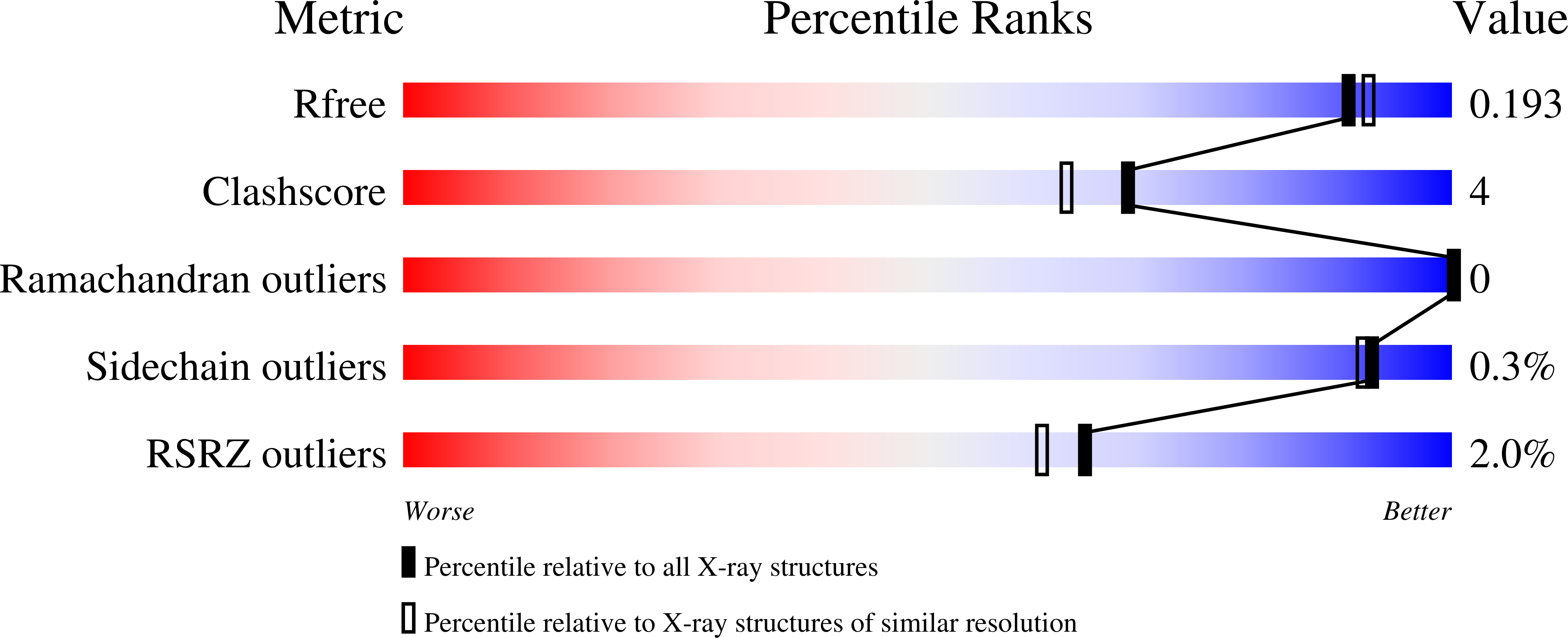
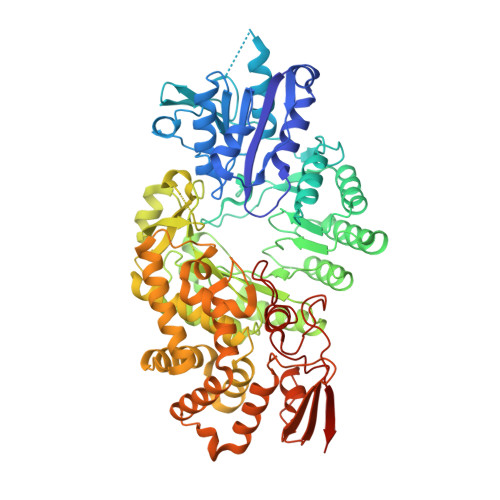
Structure of a B 12 -dependent radical SAM enzyme in carbapenem biosynthesis.
Knox, H.L., Sinner, E.K., Townsend, C.A., Boal, A.K., Booker, S.J.(2022) Nature 602: 343-348
- PubMed: 35110734
- DOI: https://doi.org/10.1038/s41586-021-04392-4
- Primary Citation of Related Structures:
7KDX, 7KDY - PubMed Abstract:
Carbapenems are antibiotics of last resort in the clinic. Owing to their potency and broad-spectrum activity, they are an important part of the antibiotic arsenal. The vital role of carbapenems is exemplified by the approval acquired by Merck from the US Food and Drug Administration (FDA) for the use of an imipenem combination therapy to treat the increased levels of hospital-acquired and ventilator-associated bacterial pneumonia that have occurred during the COVID-19 pandemic 1 . The C6 hydroxyethyl side chain distinguishes the clinically used carbapenems from the other classes of β-lactam antibiotics and is responsible for their low susceptibility to inactivation by occluding water from the β-lactamase active site 2 . The construction of the C6 hydroxyethyl side chain is mediated by cobalamin- or B 12 -dependent radical S-adenosylmethionine (SAM) enzymes 3 . These radical SAM methylases (RSMTs) assemble the alkyl backbone by sequential methylation reactions, and thereby underlie the therapeutic usefulness of clinically used carbapenems. Here we present X-ray crystal structures of TokK, a B 12 -dependent RSMT that catalyses three-sequential methylations during the biosynthesis of asparenomycin A. These structures, which contain the two metallocofactors of the enzyme and were determined in the presence and absence of a carbapenam substrate, provide a visualization of a B 12 -dependent RSMT that uses the radical mechanism that is shared by most of these enzymes. The structures provide insight into the stereochemistry of initial C6 methylation and suggest that substrate positioning governs the rate of each methylation event.
Organizational Affiliation:
Department of Chemistry, Pennsylvania State University, University Park, PA, USA.