Negative Regulation of TLR Signaling by BCAP Requires Dimerization of Its DBB Domain.
Lauenstein, J.U., Scherm, M.J., Udgata, A., Moncrieffe, M.C., Fisher, D.I., Gay, N.J.(2020) J Immunol 204: 2269-2276
- PubMed: 32198144
- DOI: https://doi.org/10.4049/jimmunol.1901210
- Primary Citation of Related Structures:
6SWS - PubMed Abstract:
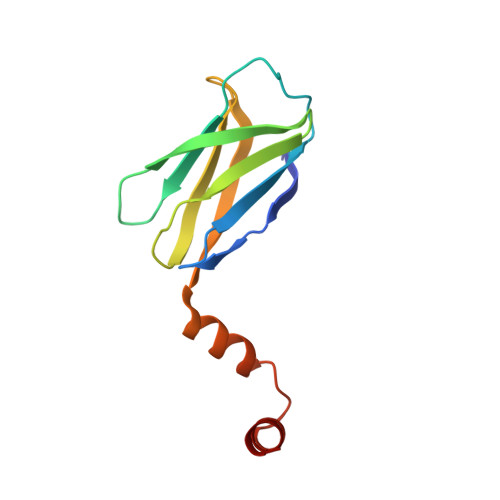
The B cell adaptor protein (BCAP) is a multimodular regulator of inflammatory signaling in diverse immune system cells. BCAP couples TLR signaling to phosphoinositide metabolism and inhibits MyD88-directed signal transduction. BCAP is recruited to the TLR signalosome forming multitypic interactions with the MAL and MyD88 signaling adaptors. In this study, we show that indirect dimerization of BCAP TIR is required for negative regulation of TLR signaling. This regulation is mediated by a transcription factor Ig (TIG/IPT) domain, a fold found in the NF-κB family of transcription factors. We have solved the crystal structure of the BCAP TIG and find that it is most similar to that of early B cell factor 1 (EBF1). In both cases, the dimer is stabilized by a helix-loop-helix motif at the C terminus and interactions between the β-sheets of the Ig domains. BCAP is exclusively localized in the cytosol and is unable to bind DNA. Thus, the TIG domain is a promiscuous dimerization module that has been appropriated for a range of regulatory functions in gene expression and signal transduction.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, Cambridge CB2 1GA, United Kingdom; and.