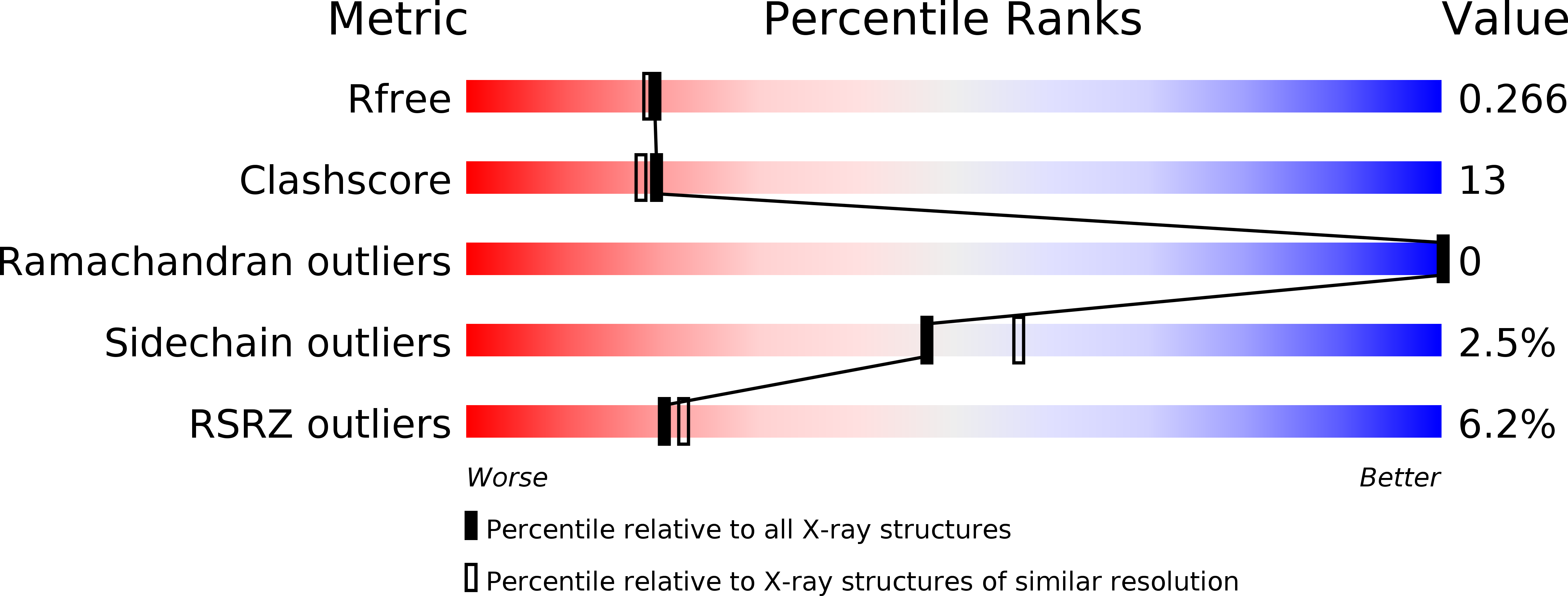
Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach.
Khusainov, I., Fatkhullin, B., Pellegrino, S., Bikmullin, A., Liu, W.T., Gabdulkhakov, A., Shebel, A.A., Golubev, A., Zeyer, D., Trachtmann, N., Sprenger, G.A., Validov, S., Usachev, K., Yusupova, G., Yusupov, M.(2020) Nat Commun 11: 1656-1656
- PubMed: 32245971
- DOI: https://doi.org/10.1038/s41467-020-15517-0
- Primary Citation of Related Structures:
6SJ5, 6SJ6 - PubMed Abstract:
For the sake of energy preservation, bacteria, upon transition to stationary phase, tone down their protein synthesis. This process is favored by the reversible binding of small stress-induced proteins to the ribosome to prevent unnecessary translation. One example is the conserved bacterial ribosome silencing factor (RsfS) that binds to uL14 protein onto the large ribosomal subunit and prevents its association with the small subunit. Here we describe the binding mode of Staphylococcus aureus RsfS to the large ribosomal subunit and present a 3.2 Å resolution cryo-EM reconstruction of the 50S-RsfS complex together with the crystal structure of uL14-RsfS complex solved at 2.3 Å resolution. The understanding of the detailed landscape of RsfS-uL14 interactions within the ribosome shed light on the mechanism of ribosome shutdown in the human pathogen S. aureus and might deliver a novel target for pharmacological drug development and treatment of bacterial infections.
Organizational Affiliation:
Laboratory of Structural Biology, Institute of Fundamental Medicine and Biology, Kazan Federal University, Kremlyovskaya Street 18, Kazan, 420008, Russia. iskander.khusainov@biophys.mpg.de.