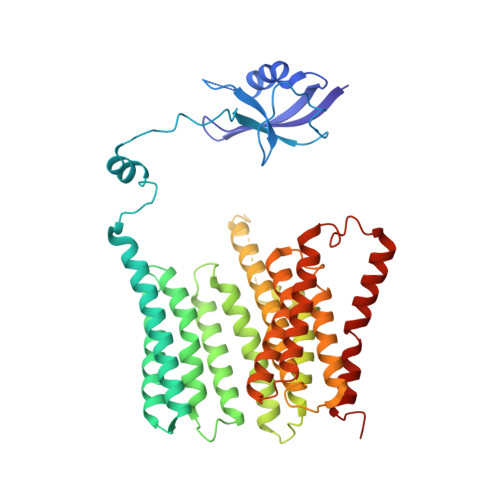
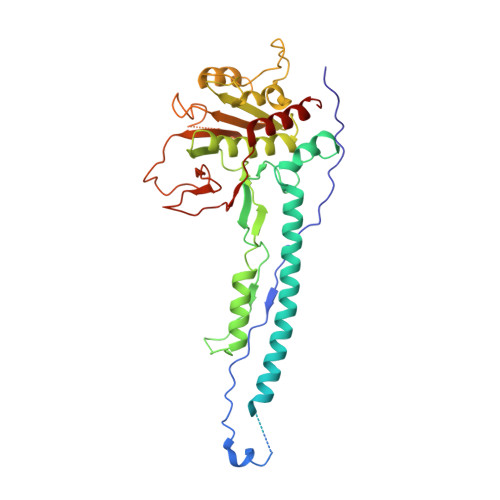
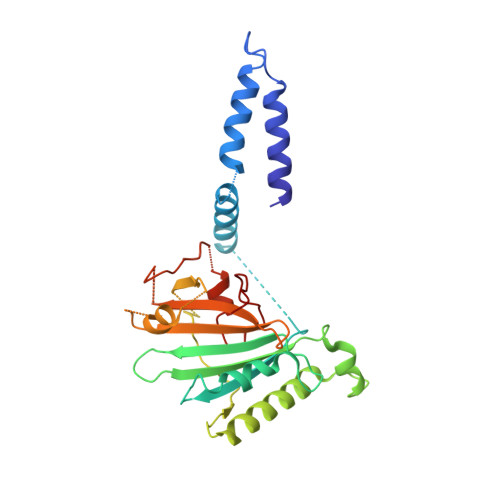
Architecture of the mycobacterial type VII secretion system.
Famelis, N., Rivera-Calzada, A., Degliesposti, G., Wingender, M., Mietrach, N., Skehel, J.M., Fernandez-Leiro, R., Bottcher, B., Schlosser, A., Llorca, O., Geibel, S.(2019) Nature 576: 321-325
- PubMed: 31597161
- DOI: https://doi.org/10.1038/s41586-019-1633-1
- Primary Citation of Related Structures:
6SGW, 6SGX, 6SGY, 6SGZ - PubMed Abstract:
Host infection by pathogenic mycobacteria, such as Mycobacterium tuberculosis, is facilitated by virulence factors that are secreted by type VII secretion systems 1 . A molecular understanding of the type VII secretion mechanism has been hampered owing to a lack of three-dimensional structures of the fully assembled secretion apparatus. Here we report the cryo-electron microscopy structure of a membrane-embedded core complex of the ESX-3/type VII secretion system from Mycobacterium smegmatis. The core of the ESX-3 secretion machine consists of four protein components-EccB3, EccC3, EccD3 and EccE3, in a 1:1:2:1 stoichiometry-which form two identical protomers. The EccC3 coupling protein comprises a flexible array of four ATPase domains, which are linked to the membrane through a stalk domain. The domain of unknown function (DUF) adjacent to the stalk is identified as an ATPase domain that is essential for secretion. EccB3 is predominantly periplasmatic, but a small segment crosses the membrane and contacts the stalk domain. This suggests that conformational changes in the stalk domain-triggered by substrate binding at the distal end of EccC3 and subsequent ATP hydrolysis in the DUF-could be coupled to substrate secretion to the periplasm. Our results reveal that the architecture of type VII secretion systems differs markedly from that of other known secretion machines 2 , and provide a structural understanding of these systems that will be useful for the design of antimicrobial strategies that target bacterial virulence.
Organizational Affiliation:
Institute for Molecular Infection Biology, Julius-Maximilians-University Würzburg, Würzburg, Germany.