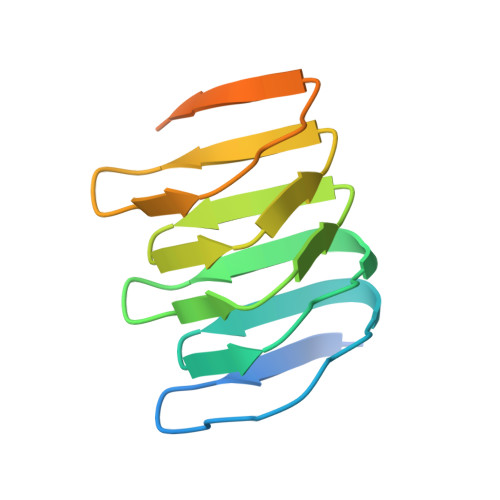
The structure of bactofilin filaments reveals their mode of membrane binding and lack of polarity.
Deng, X., Gonzalez Llamazares, A., Wagstaff, J.M., Hale, V.L., Cannone, G., McLaughlin, S.H., Kureisaite-Ciziene, D., Lowe, J.(2019) Nat Microbiol 4: 2357-2368
- PubMed: 31501539
- DOI: https://doi.org/10.1038/s41564-019-0544-0
- Primary Citation of Related Structures:
6RIA, 6RIB - PubMed Abstract:
Bactofilins are small β-helical proteins that form cytoskeletal filaments in a range of bacteria. Bactofilins have diverse functions, from cell stalk formation in Caulobacter crescentus to chromosome segregation and motility in Myxococcus xanthus. However, the precise molecular architecture of bactofilin filaments has remained unclear. Here, sequence analysis and electron microscopy results reveal that, in addition to being widely distributed across bacteria and archaea, bactofilins are also present in a few eukaryotic lineages such as the Oomycetes. Electron cryomicroscopy analysis demonstrated that the sole bactofilin from Thermus thermophilus (TtBac) forms constitutive filaments that polymerize through end-to-end association of the β-helical domains. Using a nanobody, we determined the near-atomic filament structure, showing that the filaments are non-polar. A polymerization-impairing mutation enabled crystallization and structure determination, while reaffirming the lack of polarity and the strength of the β-stacking interface. To confirm the generality of the lack of polarity, we performed coevolutionary analysis on a large set of sequences. Finally, we determined that the widely conserved N-terminal disordered tail of TtBac is responsible for direct binding to lipid membranes, both on liposomes and in Escherichia coli cells. Membrane binding is probably a common feature of these widespread but only recently discovered filaments of the prokaryotic cytoskeleton.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge, UK.