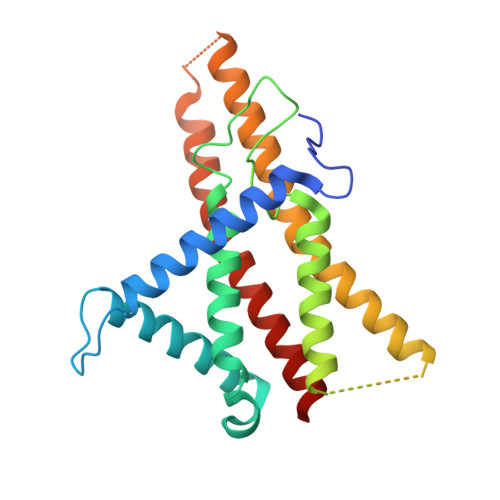
Structure of a bacterial ATP synthase.
Guo, H., Suzuki, T., Rubinstein, J.L.(2019) Elife 8
- PubMed: 30724163
- DOI: https://doi.org/10.7554/eLife.43128
- Primary Citation of Related Structures:
6N2D, 6N2Y, 6N2Z, 6N30 - PubMed Abstract:
ATP synthases produce ATP from ADP and inorganic phosphate with energy from a transmembrane proton motive force. Bacterial ATP synthases have been studied extensively because they are the simplest form of the enzyme and because of the relative ease of genetic manipulation of these complexes. We expressed the Bacillus PS3 ATP synthase in Eschericia coli , purified it, and imaged it by cryo-EM, allowing us to build atomic models of the complex in three rotational states. The position of subunit ε shows how it is able to inhibit ATP hydrolysis while allowing ATP synthesis. The architecture of the membrane region shows how the simple bacterial ATP synthase is able to perform the same core functions as the equivalent, but more complicated, mitochondrial complex. The structures reveal the path of transmembrane proton translocation and provide a model for understanding decades of biochemical analysis interrogating the roles of specific residues in the enzyme.
Organizational Affiliation:
The Hospital for Sick Children Research Institute, Toronto, Canada.