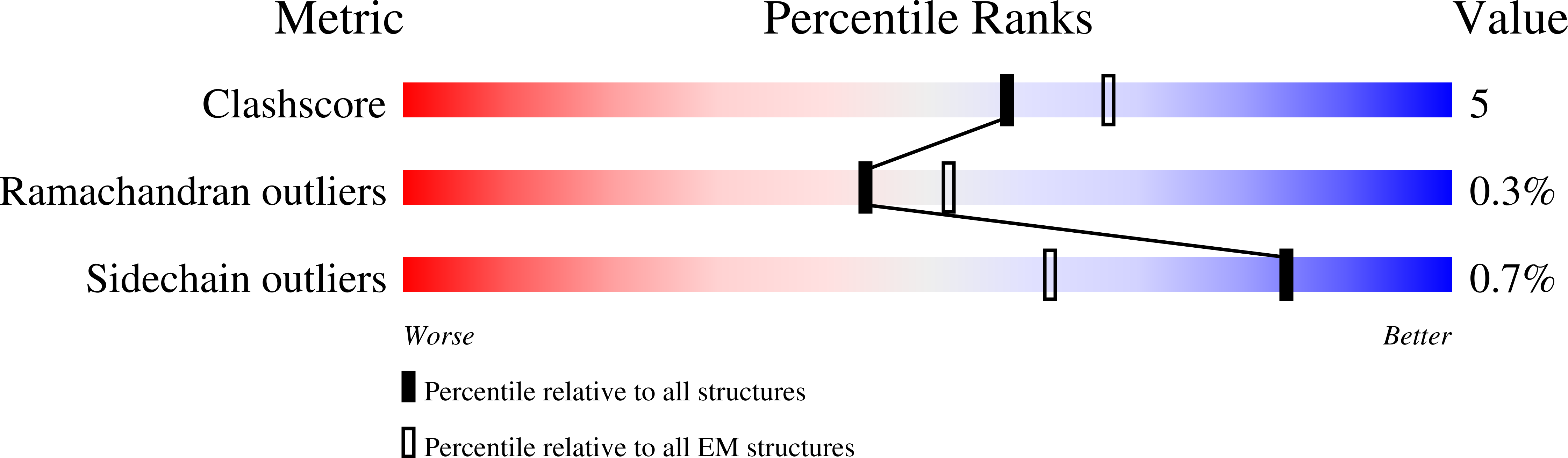Biogenesis and structure of a type VI secretion baseplate.
Cherrak, Y., Rapisarda, C., Pellarin, R., Bouvier, G., Bardiaux, B., Allain, F., Malosse, C., Rey, M., Chamot-Rooke, J., Cascales, E., Fronzes, R., Durand, E.(2018) Nat Microbiol 3: 1404-1416
- PubMed: 30323254
- DOI: https://doi.org/10.1038/s41564-018-0260-1
- Primary Citation of Related Structures:
6GIY, 6GJ1, 6GJ3 - PubMed Abstract:
To support their growth in a competitive environment and cause pathogenesis, bacteria have evolved a broad repertoire of macromolecular machineries to deliver specific effectors and toxins. Among these multiprotein complexes, the type VI secretion system (T6SS) is a contractile nanomachine that targets both prokaryotic and eukaryotic cells. The T6SS comprises two functional subcomplexes: a bacteriophage-related tail structure anchored to the cell envelope by a membrane complex. As in other contractile injection systems, the tail is composed of an inner tube wrapped by a sheath and built on the baseplate. In the T6SS, the baseplate is not only the tail assembly platform, but also docks the tail to the membrane complex and hence serves as an evolutionary adaptor. Here we define the biogenesis pathway and report the cryo-electron microscopy (cryo-EM) structure of the wedge protein complex of the T6SS from enteroaggregative Escherichia coli (EAEC). Using an integrative approach, we unveil the molecular architecture of the whole T6SS baseplate and its interaction with the tail sheath, offering detailed insights into its biogenesis and function. We discuss architectural and mechanistic similarities but also reveal key differences with the T4 phage and Mu phage baseplates.
Organizational Affiliation:
Laboratoire d'Ingénierie des Systèmes Macromoléculaires, Institut de Microbiologie de la Méditerranée, UMR7255, Aix-Marseille Université - CNRS, Marseille, France.