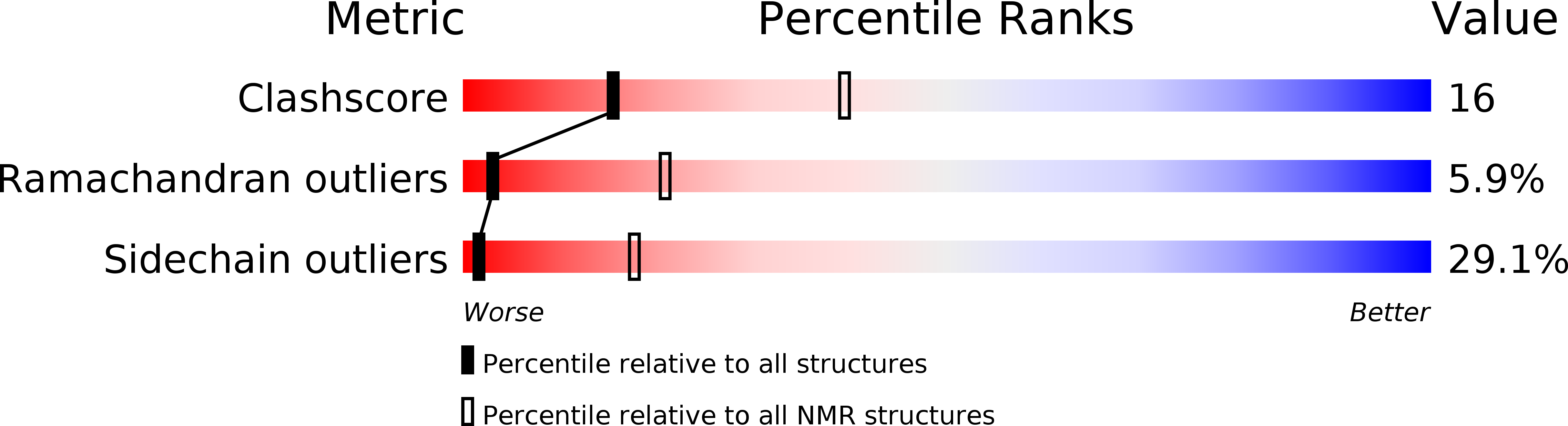Molecular Basis of Class III Ligand Recognition by PDZ3 in Murine Protein Tyrosine Phosphatase PTPN13.
Kock, G., Dicks, M., Yip, K.T., Kohl, B., Putz, S., Heumann, R., Erdmann, K.S., Stoll, R.(2018) J Mol Biol 430: 4275-4292
- PubMed: 30189200
- DOI: https://doi.org/10.1016/j.jmb.2018.08.023
- Primary Citation of Related Structures:
6GBD, 6GBE - PubMed Abstract:
Protein tyrosine phosphatase PTPN13, also known as PTP-BL in mice, represents a large multi-domain non-transmembrane scaffolding protein that contains five consecutive PDZ domains. Here, we report the solution structures of the extended murine PTPN13 PDZ3 domain in its apo form and in complex with its physiological ligand, the carboxy-terminus of protein kinase C-related kinase-2 (PRK2), determined by multidimensional NMR spectroscopy. Both in its ligand-free state and when complexed to PRK2, PDZ3 of PTPN13 adopts the classical compact, globular D/E fold. PDZ3 of PTPN13 binds five carboxy-terminal amino acids of PRK2 via a groove located between the EB-strand and the DB-helix. The PRK2 peptide resides in the canonical PDZ3 binding cleft in an elongated manner and the amino acid side chains in position P0 and P-2, cysteine and aspartate, of the ligand face the groove between EB-strand and DB-helix, whereas the PRK2 side chains of tryptophan and alanine located in position P-1 and P-3 point away from the binding cleft. These structures are rare examples of selective class III ligand recognition by a PDZ domain and now provide a basis for the detailed structural investigation of the promiscuous interaction between the PDZ domains of PTPN13 and their ligands. They will also lead to a better understanding of the proposed scaffolding function of these domains in multi-protein complexes assembled by PTPN13 and could ultimately contribute to low molecular weight antagonists that might even act on the PRK2 signaling pathway to modulate rearrangements of the actin cytoskeleton.
Organizational Affiliation:
Biomolecular NMR Spectroscopy, Faculty of Chemistry and Biochemistry, Ruhr-University of Bochum, D-44780, Germany.