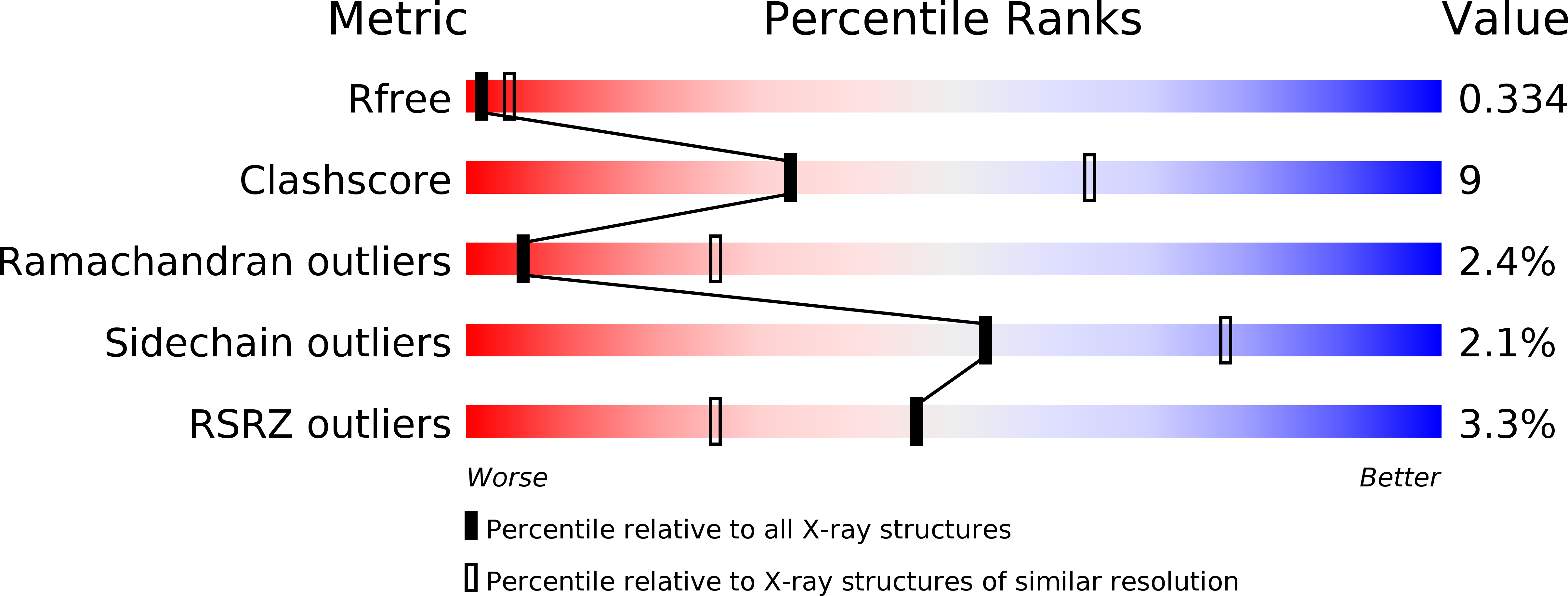
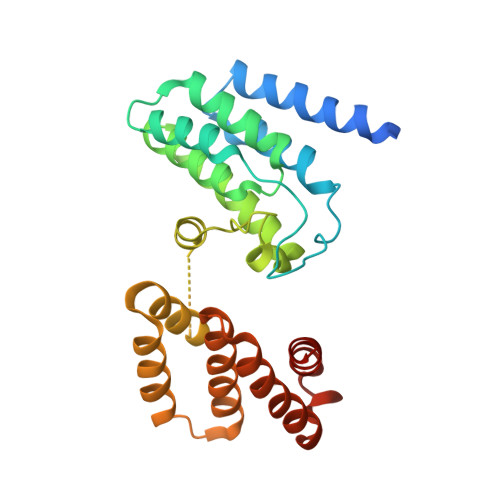
Structural insights into the function of type VI secretion system TssA subunits.
Dix, S.R., Owen, H.J., Sun, R., Ahmad, A., Shastri, S., Spiewak, H.L., Mosby, D.J., Harris, M.J., Batters, S.L., Brooker, T.A., Tzokov, S.B., Sedelnikova, S.E., Baker, P.J., Bullough, P.A., Rice, D.W., Thomas, M.S.(2018) Nat Commun 9: 4765-4765
- PubMed: 30420757
- DOI: https://doi.org/10.1038/s41467-018-07247-1
- Primary Citation of Related Structures:
6G7B, 6G7C, 6H8E, 6H8F, 6HS5, 6HS6 - PubMed Abstract:
The type VI secretion system (T6SS) is a multi-protein complex that injects bacterial effector proteins into target cells. It is composed of a cell membrane complex anchored to a contractile bacteriophage tail-like apparatus consisting of a sharpened tube that is ejected by the contraction of a sheath against a baseplate. We present structural and biochemical studies on TssA subunits from two different T6SSs that reveal radically different quaternary structures in comparison to the dodecameric E. coli TssA that arise from differences in their C-terminal sequences. Despite this, the different TssAs retain equivalent interactions with other components of the complex and position their highly conserved N-terminal ImpA_N domain at the same radius from the centre of the sheath as a result of their distinct domain architectures, which includes additional spacer domains and highly mobile interdomain linkers. Together, these variations allow these distinct TssAs to perform a similar function in the complex.
Organizational Affiliation:
Department of Molecular Biology and Biotechnology, Krebs Institute, University of Sheffield, Sheffield, S10 2TN, UK.