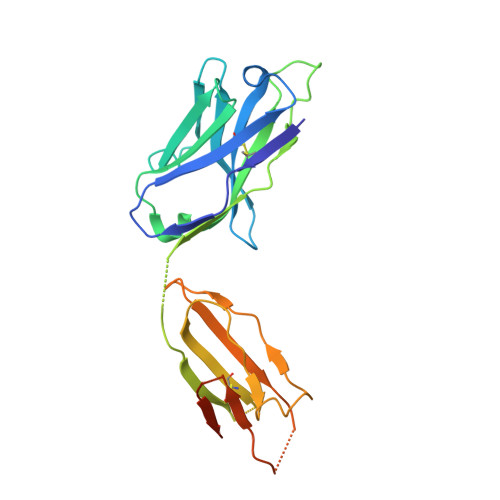
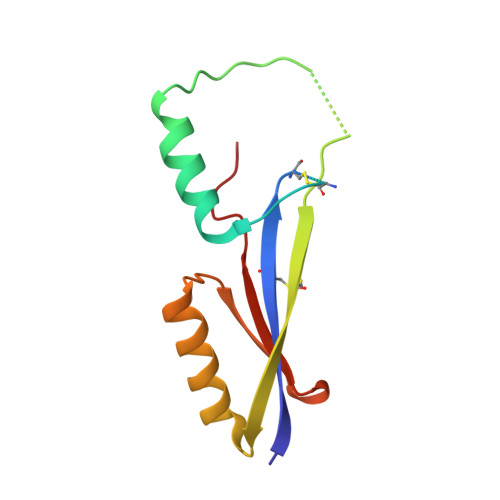
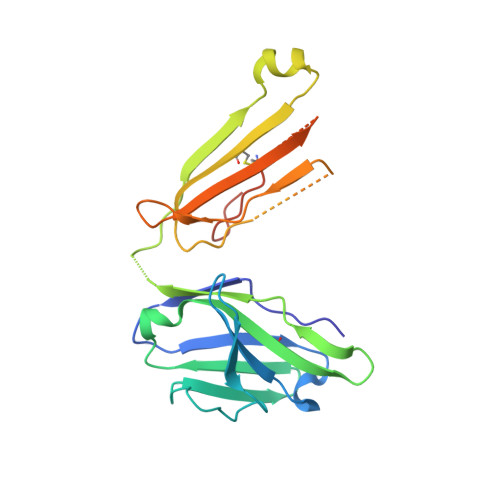
Common helical V1V2 conformations of HIV-1 Envelope expose the alpha 4 beta 7 binding site on intact virions.
Wibmer, C.K., Richardson, S.I., Yolitz, J., Cicala, C., Arthos, J., Moore, P.L., Morris, L.(2018) Nat Commun 9: 4489-4489
- PubMed: 30367034
- DOI: https://doi.org/10.1038/s41467-018-06794-x
- Primary Citation of Related Structures:
6FY1, 6FY2, 6FY3 - PubMed Abstract:
The α4β7 integrin is a non-essential HIV-1 adhesion receptor, bound by the gp120 V1V2 domain, facilitating rapid viral dissemination into gut-associated lymphoid tissues. Antibodies blocking this interaction early in infection can improve disease outcome, and V1V2-targeted antibodies were correlated with moderate efficacy reported from the RV144 HIV-1 vaccine trial. Monoclonal α4β7-blocking antibodies recognise two slightly different helical V2 conformations, and current structural data suggests their binding sites are occluded in prefusion envelope trimers. Here, we report cocrystal structures of two α4β7-blocking antibodies from an infected donor complexed with scaffolded V1V2 or V2 peptides. Both antibodies recognised the same helix-coil V2 conformation as RV144 antibody CH58, identifying a frequently sampled alternative conformation of full-length V1V2. In the context of Envelope, this α-helical form of V1V2 displays highly exposed α4β7-binding sites, potentially providing a functional role for non-native Envelope on virion or infected cell surfaces in HIV-1 dissemination, pathogenesis, and vaccine design.
Organizational Affiliation:
Centre for HIV and STIs, National Institute for Communicable Diseases (NICD), of the National Health Laboratory Service (NHLS), Johannesburg, 2131, South Africa. c.k.wibmer@gmail.com.