Identification of a peptide inhibitor for the histone methyltransferase WHSC1.
Morrison, M.J., Boriack-Sjodin, P.A., Swinger, K.K., Wigle, T.J., Sadalge, D., Kuntz, K.W., Scott, M.P., Janzen, W.P., Chesworth, R., Duncan, K.W., Harvey, D.M., Lampe, J.W., Mitchell, L.H., Copeland, R.A.(2018) PLoS One 13: e0197082-e0197082
- PubMed: 29742153
- DOI: https://doi.org/10.1371/journal.pone.0197082
- Primary Citation of Related Structures:
6CEN - PubMed Abstract:
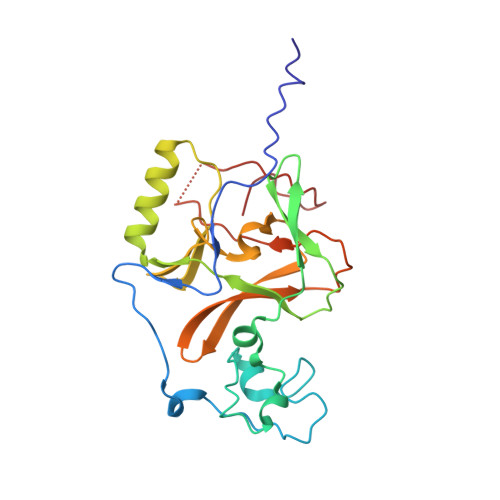
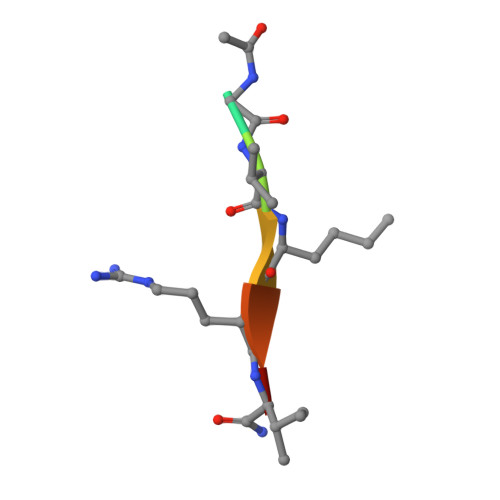
WHSC1 is a histone methyltransferase that is responsible for mono- and dimethylation of lysine 36 on histone H3 and has been implicated as a driver in a variety of hematological and solid tumors. Currently, there is a complete lack of validated chemical matter for this important drug discovery target. Herein we report on the first fully validated WHSC1 inhibitor, PTD2, a norleucine-containing peptide derived from the histone H4 sequence. This peptide exhibits micromolar affinity towards WHSC1 in biochemical and biophysical assays. Furthermore, a crystal structure was solved with the peptide in complex with SAM and the SET domain of WHSC1L1. This inhibitor is an important first step in creating potent, selective WHSC1 tool compounds for the purposes of understanding the complex biology in relation to human disease.
Organizational Affiliation:
Epizyme Inc., Cambridge, Massachusetts, United States of America.