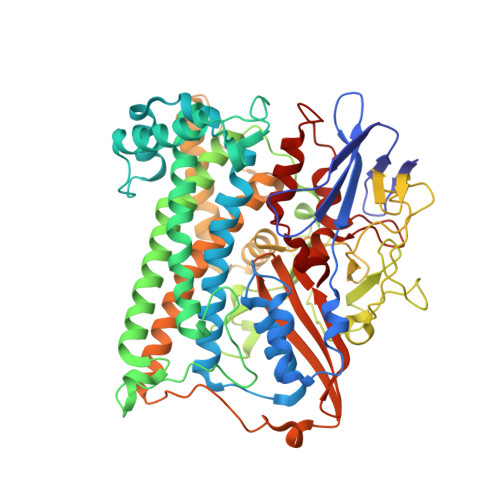
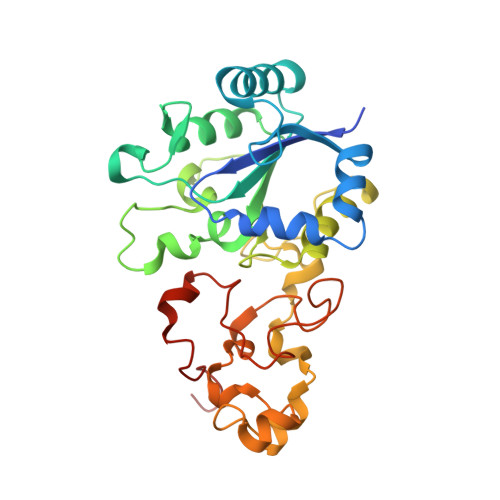
Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Shomura, Y., Yoon, K.S., Nishihara, H., Higuchi, Y.(2011) Nature 479: 253-256
- PubMed: 22002607
- DOI: https://doi.org/10.1038/nature10504
- Primary Citation of Related Structures:
3AYX, 3AYZ, 5Y34 - PubMed Abstract:
Membrane-bound respiratory [NiFe]-hydrogenase (MBH), a H(2)-uptake enzyme found in the periplasmic space of bacteria, catalyses the oxidation of dihydrogen: H(2) → 2H(+) + 2e(-) (ref. 1). In contrast to the well-studied O(2)-sensitive [NiFe]-hydrogenases (referred to as the standard enzymes), MBH has an O(2)-tolerant H(2) oxidation activity; however, the mechanism of O(2) tolerance is unclear. Here we report the crystal structures of Hydrogenovibrio marinus MBH in three different redox conditions at resolutions between 1.18 and 1.32 Å. We find that the proximal iron-sulphur (Fe-S) cluster of MBH has a [4Fe-3S] structure coordinated by six cysteine residues--in contrast to the [4Fe-4S] cubane structure coordinated by four cysteine residues found in the proximal Fe-S cluster of the standard enzymes--and that an amide nitrogen of the polypeptide backbone is deprotonated and additionally coordinates the cluster when chemically oxidized, thus stabilizing the superoxidized state of the cluster. The structure of MBH is very similar to that of the O(2)-sensitive standard enzymes except for the proximal Fe-S cluster. Our results give a reasonable explanation why the O(2) tolerance of MBH is attributable to the unique proximal Fe-S cluster; we propose that the cluster is not only a component of the electron transfer for the catalytic cycle, but that it also donates two electrons and one proton crucial for the appropriate reduction of O(2) in preventing the formation of an unready, inactive state of the enzyme.
Organizational Affiliation:
Department of Life Science, Graduate School of Life Science, University of Hyogo, 3-2-1 Koto, Kamigori-cho, Ako-gun, Hyogo 678-1297, Japan.