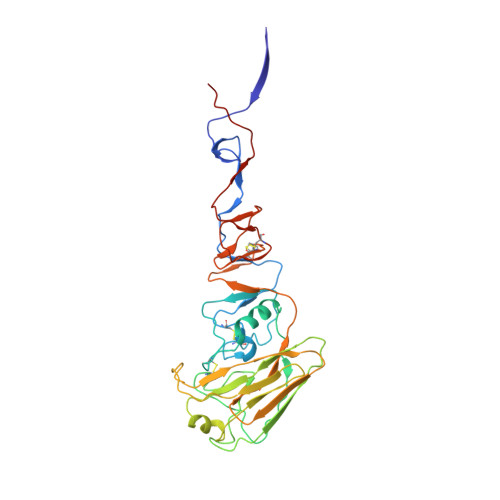
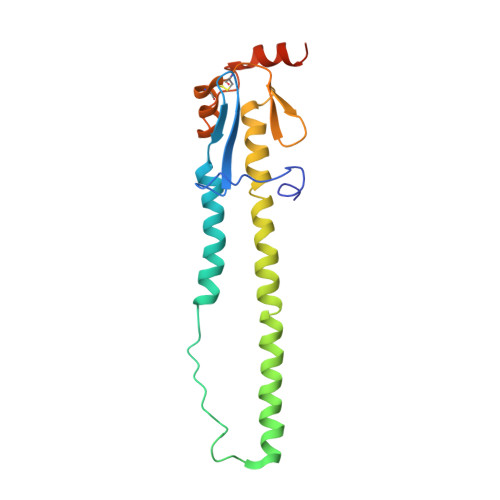
H7N9 influenza virus neutralizing antibodies that possess few somatic mutations.
Thornburg, N.J., Zhang, H., Bangaru, S., Sapparapu, G., Kose, N., Lampley, R.M., Bombardi, R.G., Yu, Y., Graham, S., Branchizio, A., Yoder, S.M., Rock, M.T., Creech, C.B., Edwards, K.M., Lee, D., Li, S., Wilson, I.A., Garcia-Sastre, A., Albrecht, R.A., Crowe, J.E.(2016) J Clin Invest 126: 1482-1494
- PubMed: 26950424
- DOI: https://doi.org/10.1172/JCI85317
- Primary Citation of Related Structures:
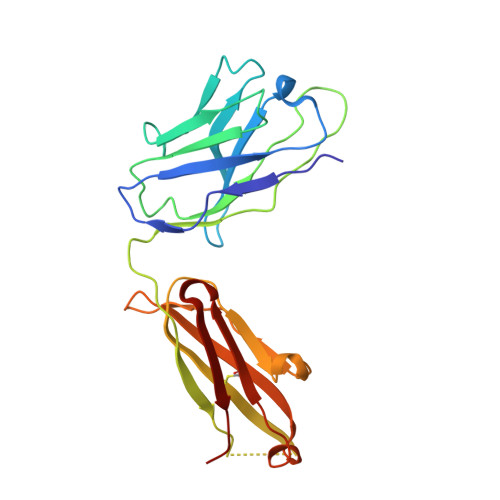
5V2A - PubMed Abstract:
Avian H7N9 influenza viruses are group 2 influenza A viruses that have been identified as the etiologic agent for a current major outbreak that began in China in 2013 and may pose a pandemic threat. Here, we examined the human H7-reactive antibody response in 75 recipients of a monovalent inactivated A/Shanghai/02/2013 H7N9 vaccine. After 2 doses of vaccine, the majority of donors had memory B cells that secreted IgGs specific for H7 HA, with dominant responses against single HA subtypes, although frequencies of H7-reactive B cells ranged widely between donors. We isolated 12 naturally occurring mAbs with low half-maximal effective concentrations for binding, 5 of which possessed neutralizing and HA-inhibiting activities. The 5 neutralizing mAbs exhibited narrow breadth of reactivity with influenza H7 strains. Epitope-mapping studies using neutralization escape mutant analysis, deuterium exchange mass spectrometry, and x-ray crystallography revealed that these neutralizing mAbs bind near the receptor-binding pocket on HA. All 5 neutralizing mAbs possessed low numbers of somatic mutations, suggesting the clones arose from naive B cells. The most potent mAb, H7.167, was tested as a prophylactic treatment in a mouse intranasal virus challenge study, and systemic administration of the mAb markedly reduced viral lung titers.