Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Granell, M., Namura, M., Alvira, S., Kanamaru, S., van Raaij, M.J.(2017) Viruses 9
- PubMed: 28665339
- DOI: https://doi.org/10.3390/v9070168
- Primary Citation of Related Structures:
4UXE, 4UXF, 4UXG, 5NXF, 5NXH - PubMed Abstract:
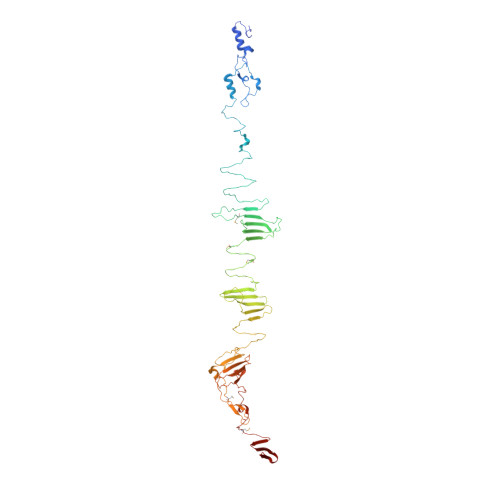
Long tail fibers of bacteriophage T4 are formed by proteins gp34, gp35, gp36, and gp37, with gp34 located at the phage-proximal end and gp37 at the phage-distal, receptor-binding end. We have solved the structure of the carboxy-terminal region of gp34, consisting of amino acids 894-1289, by single-wavelength anomalous diffraction and extended the structure to amino acids 744-1289 using data collected from crystals containing longer gp34-fragments. The structure reveals three repeats of a mixed α-β fibrous domain in residues 744 to 877. A triple-helical neck connects to an extended triple β-helix domain (amino acids 900-1127) punctuated by two β-prism domains. Next, a β-prism domain decorated with short helices and extended β-helices is present (residues 1146-1238), while the C -terminal end is capped with another short β-helical region and three β-hairpins. The structure provides insight into the stability of the fibrous gp34 protein.
Organizational Affiliation:
Departmento de Estructura de Macromoleculas, Centro Nacional de Biotecnologia (CNB-CSIC), Calle Darwin 3, E-28049 Madrid, Spain.