Conformational Plasticity in the Transsynaptic Neurexin-Cerebellin-Glutamate Receptor Adhesion Complex.
Cheng, S., Seven, A.B., Wang, J., Skiniotis, G., Ozkan, E.(2016) Structure 24: 2163-2173
- PubMed: 27926833
- DOI: https://doi.org/10.1016/j.str.2016.11.004
- Primary Citation of Related Structures:
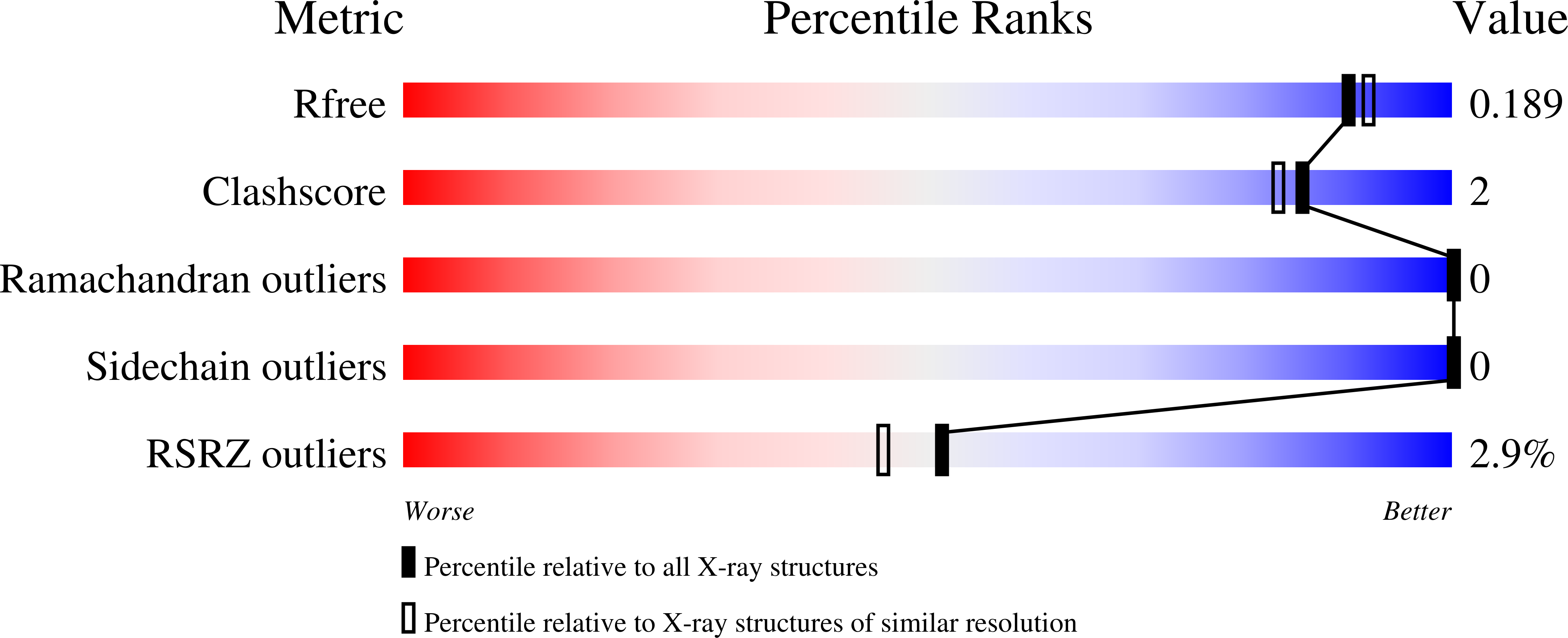
5KWR, 5L2E - PubMed Abstract:
Synaptic specificity is a defining property of neural networks. In the cerebellum, synapses between parallel fiber neurons and Purkinje cells are specified by the simultaneous interactions of secreted protein cerebellin with pre-synaptic neurexin and post-synaptic delta-type glutamate receptors (GluD). Here, we determined the crystal structures of the trimeric C1q-like domain of rat cerebellin-1, and the first complete ectodomain of a GluD, rat GluD2. Cerebellin binds to the LNS6 domain of α- and β-neurexin-1 through a high-affinity interaction that involves its highly flexible N-terminal domain. In contrast, we show that the interaction of cerebellin with isolated GluD2 ectodomain is low affinity, which is not simply an outcome of lost avidity when compared with binding with a tetrameric full-length receptor. Rather, high-affinity capture of cerebellin by post-synaptic terminals is likely controlled by long-distance regulation within this transsynaptic complex. Altogether, our results suggest unusual conformational flexibility within all components of the complex.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, IL 60637, USA.