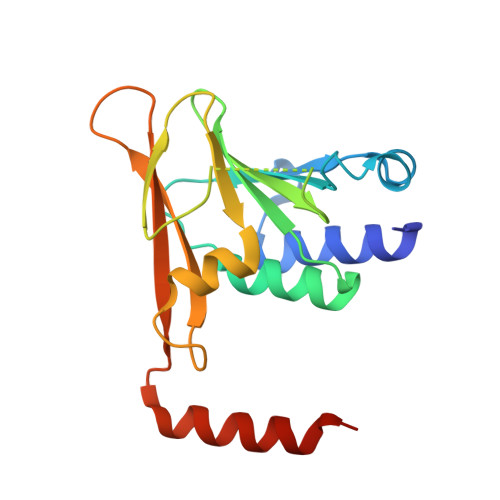
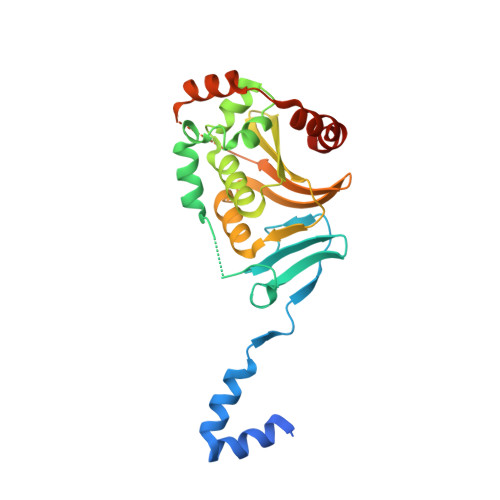
Crystal Structure of the Human Cytomegalovirus pUL50-pUL53 Core Nuclear Egress Complex Provides Insight into a Unique Assembly Scaffold for Virus-Host Protein Interactions.
Walzer, S.A., Egerer-Sieber, C., Sticht, H., Sevvana, M., Hohl, K., Milbradt, J., Muller, Y.A., Marschall, M.(2015) J Biol Chem 290: 27452-27458
- PubMed: 26432641
- DOI: https://doi.org/10.1074/jbc.C115.686527
- Primary Citation of Related Structures:
5D5N - PubMed Abstract:
Nuclear replication of cytomegalovirus relies on elaborate mechanisms of nucleocytoplasmic egress of viral particles. Thus, the role of two essential and conserved viral nuclear egress proteins, pUL50 and pUL53, is pivotal. pUL50 and pUL53 heterodimerize and form a core nuclear egress complex (NEC), which is anchored to the inner nuclear membrane and provides a scaffold for the assembly of a multimeric viral-cellular NEC. Here, we report the crystal structure of the pUL50-pUL53 heterodimer (amino acids 1-175 and 50-292, respectively) at 2.44 Å resolution. Both proteins adopt a globular fold with mixed α and β secondary structure elements. pUL53-specific features include a zinc-binding site and a hook-like N-terminal extension, the latter representing a hallmark element of the pUL50-pUL53 interaction. The hook-like extension (amino acids 59-87) embraces pUL50 and contributes 1510 Å(2) to the total interface area (1880 Å(2)). The pUL50 structure overall resembles the recently published NMR structure of the murine cytomegalovirus homolog pM50 but reveals a considerable repositioning of the very C-terminal α-helix of pUL50 upon pUL53 binding. pUL53 shows structural resemblance with the GHKL domain of bacterial sensory histidine kinases. A close examination of the crystal structure indicates partial assembly of pUL50-pUL53 heterodimers to hexameric ring-like structures possibly providing additional scaffolding opportunities for NEC. In combination, the structural information on pUL50-pUL53 considerably improves our understanding of the mechanism of HCMV nuclear egress. It may also accelerate the validation of the NEC as a unique target for developing a novel type of antiviral drug and improved options of broad-spectrum antiherpesviral therapy.
Organizational Affiliation:
From the Division of Biotechnology, Department of Biology.