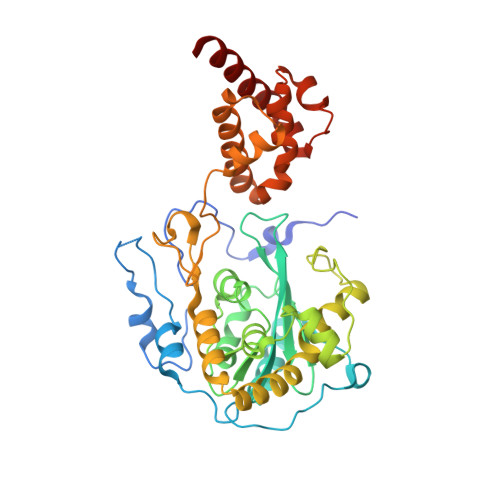
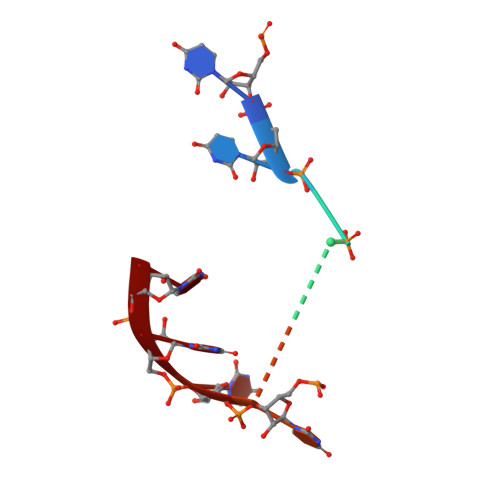
RNA degradation paths in a 12-subunit nuclear exosome complex.
Makino, D.L., Schuch, B., Stegmann, E., Baumgartner, M., Basquin, C., Conti, E.(2015) Nature 524: 54-58
- PubMed: 26222026
- DOI: https://doi.org/10.1038/nature14865
- Primary Citation of Related Structures:
5C0W, 5C0X, 5C0Y - PubMed Abstract:
The eukaryotic exosome is a conserved RNA-degrading complex that functions in RNA surveillance, turnover and processing. How the same machinery can either completely degrade or precisely trim RNA substrates has long remained unexplained. Here we report the crystal structures of a yeast nuclear exosome containing the 9-subunit core, the 3'-5' RNases Rrp44 and Rrp6, and the obligate Rrp6-binding partner Rrp47 in complex with different RNAs. The combined structural and biochemical data of this 12-subunit complex reveal how a single-stranded RNA can reach the Rrp44 or Rrp6 active sites directly or can bind Rrp6 and be threaded via the central channel towards the distal RNase Rrp44. When a bulky RNA is stalled at the entrance of the channel, Rrp6-Rrp47 swings open. The results suggest how the same molecular machine can coordinate processive degradation and partial trimming in an RNA-dependent manner by a concerted swinging mechanism of the two RNase subunits.
Organizational Affiliation:
Department of Structural Cell Biology, Max Planck Institute of Biochemistry, 82152 Martinsried, Germany.