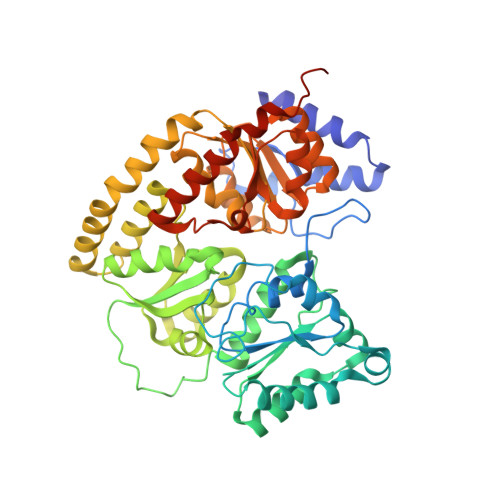
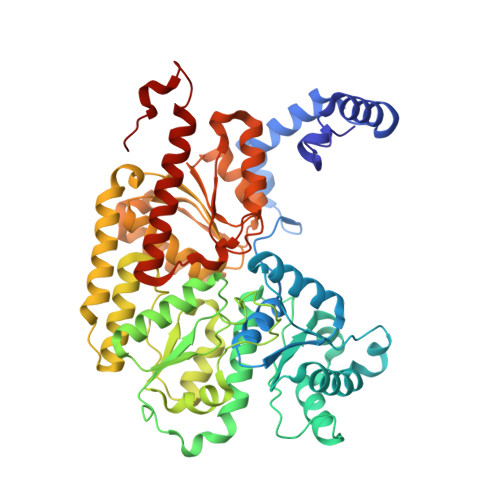
Catalysis-dependent selenium incorporation and migration in the nitrogenase active site iron-molybdenum cofactor.
Spatzal, T., Perez, K.A., Howard, J.B., Rees, D.C.(2015) Elife 4: e11620-e11620
- PubMed: 26673079
- DOI: https://doi.org/10.7554/eLife.11620
- Primary Citation of Related Structures:
5BVG, 5BVH - PubMed Abstract:
Dinitrogen reduction in the biological nitrogen cycle is catalyzed by nitrogenase, a two-component metalloenzyme. Understanding of the transformation of the inert resting state of the active site FeMo-cofactor into an activated state capable of reducing dinitrogen remains elusive. Here we report the catalysis dependent, site-selective incorporation of selenium into the FeMo-cofactor from selenocyanate as a newly identified substrate and inhibitor. The 1.60 Å resolution structure reveals selenium occupying the S2B site of FeMo-cofactor in the Azotobacter vinelandii MoFe-protein, a position that was recently identified as the CO-binding site. The Se2B-labeled enzyme retains substrate reduction activity and marks the starting point for a crystallographic pulse-chase experiment of the active site during turnover. Through a series of crystal structures obtained at resolutions of 1.32-1.66 Å, including the CO-inhibited form of Av1-Se2B, the exchangeability of all three belt-sulfur sites is demonstrated, providing direct insights into unforeseen rearrangements of the metal center during catalysis.
Organizational Affiliation:
Howard Hughes Medical Institute, California Institute of Technology, Pasadena, United States.