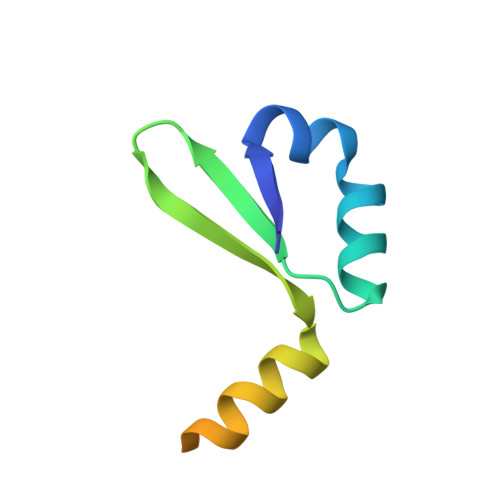
An intrinsically disordered entropic switch determines allostery in Phd-Doc regulation.
Garcia-Pino, A., De Gieter, S., Talavera, A., De Greve, H., Efremov, R.G., Loris, R.(2016) Nat Chem Biol 12: 490-496
- PubMed: 27159580
- DOI: https://doi.org/10.1038/nchembio.2078
- Primary Citation of Related Structures:
4ZLX, 4ZM0, 4ZM2 - PubMed Abstract:
Conditional cooperativity is a common mechanism involved in transcriptional regulation of prokaryotic type II toxin-antitoxin operons and is intricately related to bacterial persistence. It allows the toxin component of a toxin-antitoxin module to act as a co-repressor at low doses of toxin as compared to antitoxin. When toxin level exceeds a certain threshold, however, the toxin becomes a de-repressor. Most antitoxins contain an intrinsically disordered region (IDR) that typically is involved in toxin neutralization and repressor complex formation. To address how the antitoxin IDR is involved in transcription regulation, we studied the phd-doc operon from bacteriophage P1. We provide evidence that the IDR of Phd provides an entropic barrier precluding full operon repression in the absence of Doc. Binding of Doc results in a cooperativity switch and consequent strong operon repression, enabling context-specific modulation of the regulatory process. Variations of this theme are likely to be a common mechanism in the autoregulation of bacterial operons that involve intrinsically disordered regions.
Organizational Affiliation:
Structural Biology Brussels, Department of Biotechnology, Vrije Universiteit Brussel (VUB), Brussels, Belgium.