Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Tailford, L.E., Owen, C.D., Walshaw, J., Crost, E.H., Hardy-Goddard, J., Le Gall, G., de Vos, W.M., Taylor, G.L., Juge, N.(2015) Nat Commun 6: 7624-7624
- PubMed: 26154892
- DOI: https://doi.org/10.1038/ncomms8624
- Primary Citation of Related Structures:
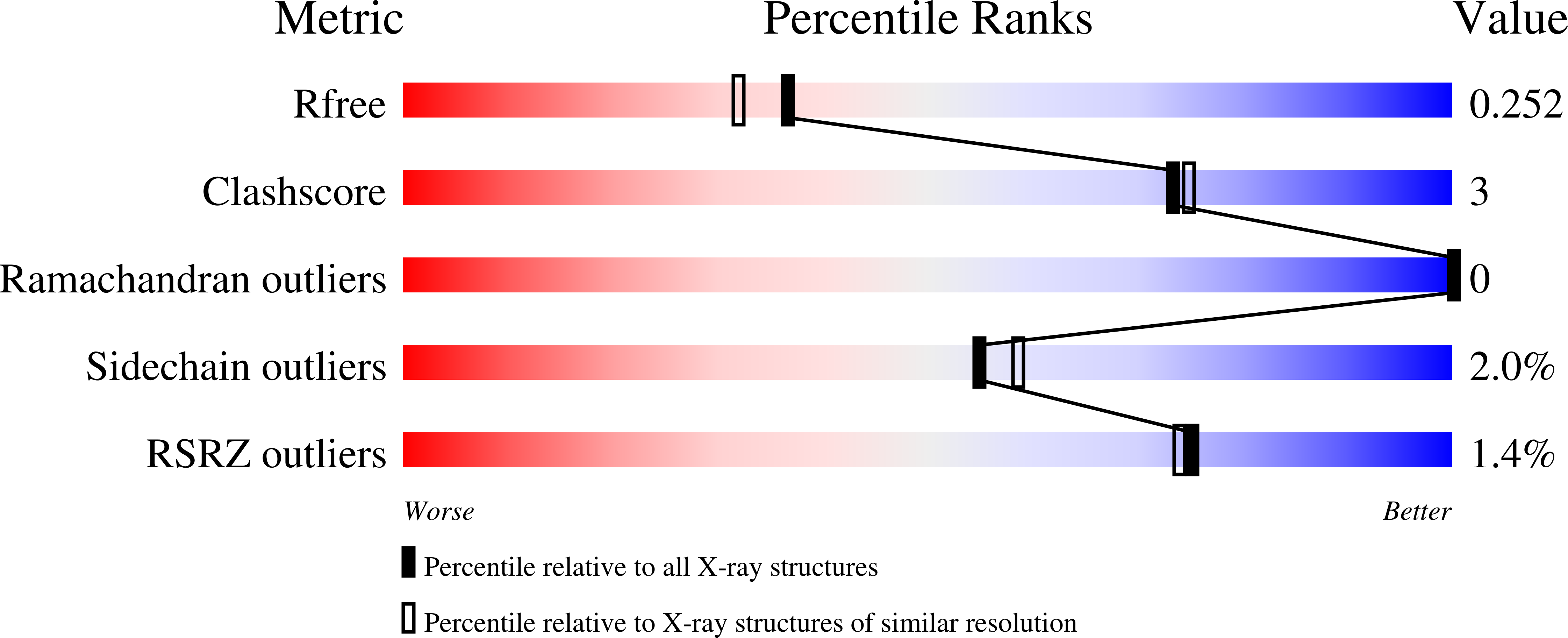
4X47, 4X49, 4X4A, 4X6K - PubMed Abstract:
The gastrointestinal mucus layer is colonized by a dense community of microbes catabolizing dietary and host carbohydrates during their expansion in the gut. Alterations in mucosal carbohydrate availability impact on the composition of microbial species. Ruminococcus gnavus is a commensal anaerobe present in the gastrointestinal tract of >90% of humans and overrepresented in inflammatory bowel diseases (IBD). Using a combination of genomics, enzymology and crystallography, we show that the mucin-degrader R. gnavus ATCC 29149 strain produces an intramolecular trans-sialidase (IT-sialidase) that cleaves off terminal α2-3-linked sialic acid from glycoproteins, releasing 2,7-anhydro-Neu5Ac instead of sialic acid. Evidence of IT-sialidases in human metagenomes indicates that this enzyme occurs in healthy subjects but is more prevalent in IBD metagenomes. Our results uncover a previously unrecognized enzymatic activity in the gut microbiota, which may contribute to the adaptation of intestinal bacteria to the mucosal environment in health and disease.
Organizational Affiliation:
The Gut Health and Food Safety Institute Strategic Programme, Institute of Food Research, Norwich Research Park, Norwich NR4 7UA, UK.