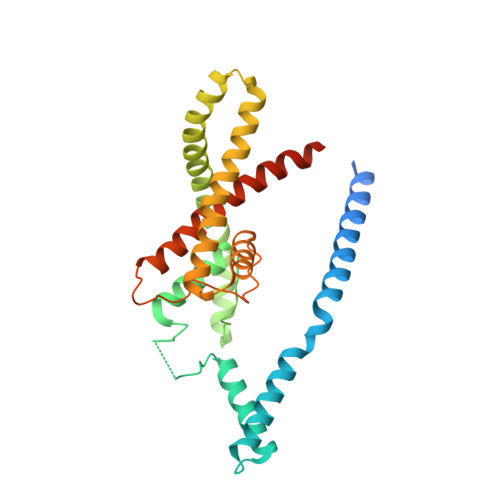
Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel.
Brohawn, S.G., Campbell, E.B., MacKinnon, R.(2014) Nature 516: 126-130
- PubMed: 25471887
- DOI: https://doi.org/10.1038/nature14013
- Primary Citation of Related Structures:
4WFE, 4WFF, 4WFG, 4WFH - PubMed Abstract:
Activation of mechanosensitive ion channels by physical force underlies many physiological processes including the sensation of touch, hearing and pain. TRAAK (also known as KCNK4) ion channels are neuronally expressed members of the two-pore domain K(+) (K2P) channel family and are mechanosensitive. They are involved in controlling mechanical and temperature nociception in mice. Mechanosensitivity of TRAAK is mediated directly through the lipid bilayer--it is a membrane-tension-gated channel. However, the molecular mechanism of TRAAK channel gating and mechanosensitivity is unknown. Here we present crystal structures of TRAAK in conductive and non-conductive conformations defined by the presence of permeant ions along the conduction pathway. In the non-conductive state, a lipid acyl chain accesses the channel cavity through a 5 Å-wide lateral opening in the membrane inner leaflet and physically blocks ion passage. In the conductive state, rotation of a transmembrane helix (TM4) about a central hinge seals the intramembrane opening, preventing lipid block of the cavity and permitting ion entry. Additional rotation of a membrane interacting TM2-TM3 segment, unique to mechanosensitive K2Ps, against TM4 may further stabilize the conductive conformation. Comparison of the structures reveals a biophysical explanation for TRAAK mechanosensitivity--an expansion in cross-sectional area up to 2.7 nm(2) in the conductive state is expected to create a membrane-tension-dependent energy difference between conformations that promotes force activation. Our results show how tension of the lipid bilayer can be harnessed to control gating and mechanosensitivity of a eukaryotic ion channel.
Organizational Affiliation:
Laboratory of Molecular Neurobiology and Biophysics and Howard Hughes Medical Institute, The Rockefeller University, 1230 York Avenue, New York, New York 10065, USA.