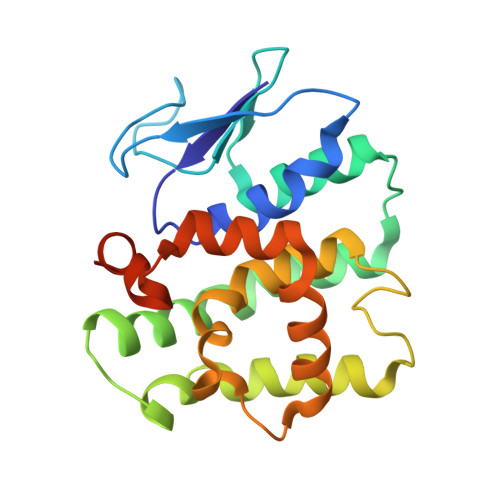
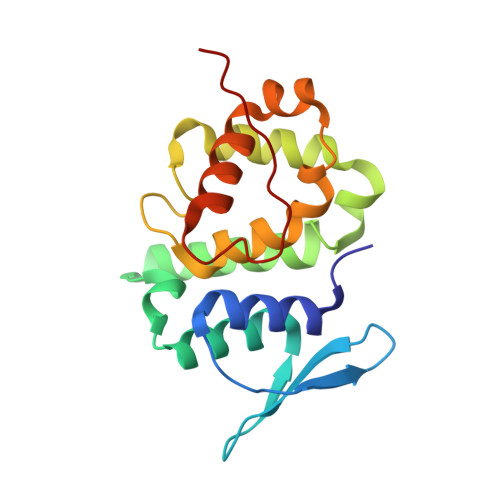
Assembly of Multi-tRNA Synthetase Complex Via Heterotetrameric Glutathione Transferase-Homology Domains.
Cho, H.Y., Maeng, S.J., Cho, H.J., Choi, Y.S., Chung, J.M., Lee, S., Kim, H.K., Kim, J.H., Eom, C., Kim, Y., Guo, M., Jung, H.S., Kang, B.S., Kim, S.(2015) J Biol Chem 290: 29313
- PubMed: 26472928
- DOI: https://doi.org/10.1074/jbc.M115.690867
- Primary Citation of Related Structures:
4BVX, 5A34, 5BMU - PubMed Abstract:
Many multicomponent protein complexes mediating diverse cellular processes are assembled through scaffolds with specialized protein interaction modules. The multi-tRNA synthetase complex (MSC), consisting of nine different aminoacyl-tRNA synthetases and three non-enzymatic factors (AIMP1-3), serves as a hub for many signaling pathways in addition to its role in protein synthesis. However, the assembly process and structural arrangement of the MSC components are not well understood. Here we show the heterotetrameric complex structure of the glutathione transferase (GST) domains shared among the four MSC components, methionyl-tRNA synthetase (MRS), glutaminyl-prolyl-tRNA synthetase (EPRS), AIMP2 and AIMP3. The MRS-AIMP3 and EPRS-AIMP2 using interface 1 are bridged via interface 2 of AIMP3 and EPRS to generate a unique linear complex of MRS-AIMP3:EPRS-AIMP2 at the molar ratio of (1:1):(1:1). Interestingly, the affinity at interface 2 of AIMP3:EPRS can be varied depending on the occupancy of interface 1, suggesting the dynamic nature of the linear GST tetramer. The four components are optimally arranged for maximal accommodation of additional domains and proteins. These characteristics suggest the GST tetramer as a unique and dynamic structural platform from which the MSC components are assembled. Considering prevalence of the GST-like domains, this tetramer can also provide a tool for the communication of the MSC with other GST-containing cellular factors.
Organizational Affiliation:
From the School of Life Science and Biotechnology, KNU Creative BioResearch Group, Kyungpook National University, Daegu 702-701, Korea.