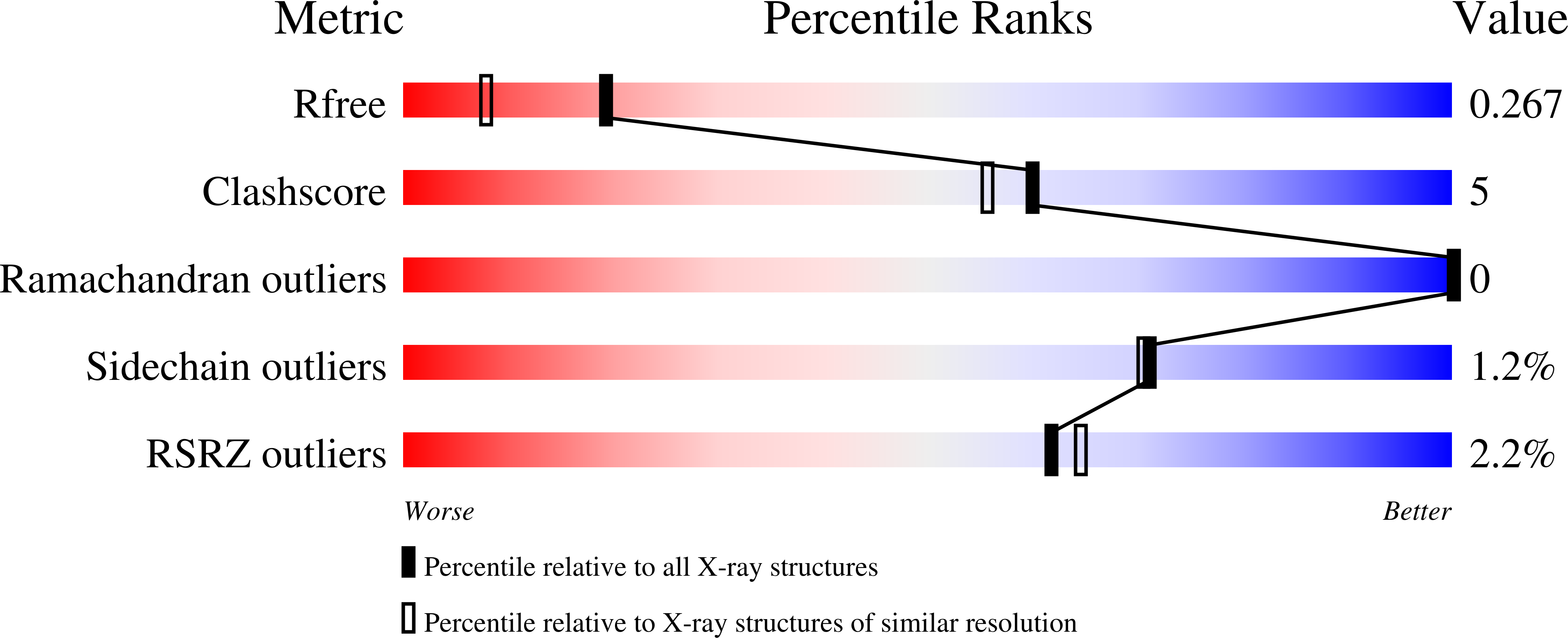
Bacteriophage T5 Encodes a Homolog of the Eukaryotic Transcription Coactivator Pc4 Implicated in Recombination-Dependent DNA Replication.
Steigemann, B., Schulz, A., Werten, S.(2013) J Mol Biol 425: 4125
- PubMed: 24029071
- DOI: https://doi.org/10.1016/j.jmb.2013.09.001
- Primary Citation of Related Structures:
4BG7 - PubMed Abstract:
The RNA polymerase II cofactor PC4 globally regulates transcription of protein-encoding genes through interactions with unwinding DNA, the basal transcription machinery and transcription activators. Here, we report the surprising identification of PC4 homologs in all sequenced representatives of the T5 family of bacteriophages, as well as in an archaeon and seven phyla of eubacteria. We have solved the crystal structure of the full-length T5 protein at 1.9Å, revealing a striking resemblance to the characteristic single-stranded DNA (ssDNA)-binding core domain of PC4. Intriguing novel structural features include a potential regulatory region at the N-terminus and a C-terminal extension of the homodimerisation interface. The genome organisation of T5-related bacteriophages points at involvement of the PC4 homolog in recombination-dependent DNA replication, strongly suggesting that the protein corresponds to the hitherto elusive replicative ssDNA-binding protein of the T5 family. Our findings imply that PC4-like factors intervene in multiple unwinding-related processes by acting as versatile modifiers of nucleic acid conformation and raise the possibility that the eukaryotic transcription coactivator derives from ancestral DNA replication, recombination and repair factors.
Organizational Affiliation:
Institute for Biochemistry, University of Greifswald, Felix-Hausdorff-Strasse 4, D-17487 Greifswald, Germany.