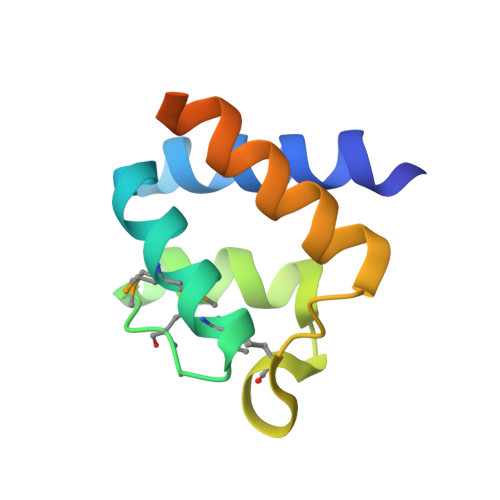
Structural analysis of HopPmaL reveals the presence of a second adaptor domain common to the HopAB family of Pseudomonas syringae type III effectors.
Singer, A.U., Wu, B., Yee, A., Houliston, S., Xu, X., Cui, H., Skarina, T., Garcia, M., Semesi, A., Arrowsmith, C.H., Savchenko, A.(2012) Biochemistry 51: 1-3
- PubMed: 22191472
- DOI: https://doi.org/10.1021/bi2013883
- Primary Citation of Related Structures:
2LF3, 2LF6, 3SVI, 3TJY - PubMed Abstract:
HopPmaL is a member of the HopAB family of type III effectors present in the phytopathogen Pseudomonas syringae. Using both X-ray crystallography and solution nuclear magnetic resonance, we demonstrate that HopPmaL contains two structurally homologous yet functionally distinct domains. The N-terminal domain corresponds to the previously described Pto-binding domain, while the previously uncharacterised C-terminal domain spans residues 308-385. While structurally similar, these domains do not share significant sequence similarity and most importantly demonstrate significant differences in key residues involved in host protein recognition, suggesting that each of them targets a different host protein.
Organizational Affiliation:
Department of Chemical Engineering and Applied Chemistry, Banting and Best Department of Medical Research, University of Toronto, Toronto, Ontario M5G 1L6, Canada.