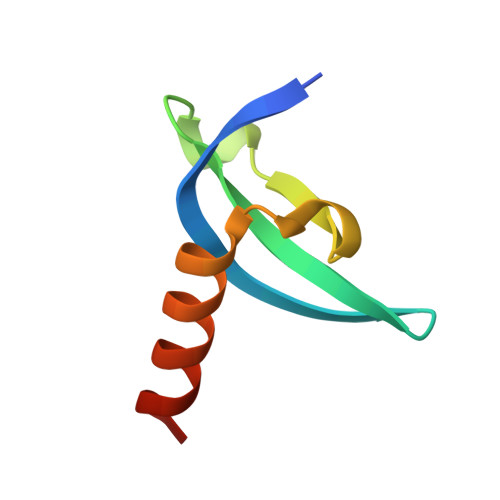
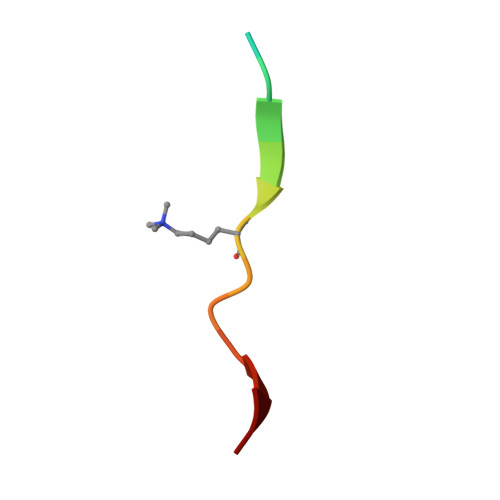
Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Li, J., Li, Z., Ruan, J., Xu, C., Tong, Y., Pan, P.W., Tempel, W., Crombet, L., Min, J., Zang, J.(2011) PLoS One 6: e25104-e25104
- PubMed: 22022377
- DOI: https://doi.org/10.1371/journal.pone.0025104
- Primary Citation of Related Structures:
3LWE, 3R93 - PubMed Abstract:
M-phase phosphoprotein 8 (MPP8) was initially identified to be a component of the RanBPM-containing large protein complex, and has recently been shown to bind to methylated H3K9 both in vivo and in vitro. MPP8 binding to methylated H3K9 is suggested to recruit the H3K9 methyltransferases GLP and ESET, and DNA methyltransferase 3A to the promoter of the E-cadherin gene, mediating the E-cadherin gene silencing and promote tumor cell motility and invasion. MPP8 contains a chromodomain in its N-terminus, which is used to bind the methylated H3K9. Here, we reported the crystal structures of human MPP8 chromodomain alone and in complex with the trimethylated histone H3K9 peptide (residue 1-15). The complex structure unveils that the human MPP8 chromodomain binds methylated H3K9 through a conserved recognition mechanism, which was also observed in Drosophila HP1, a chromodomain containing protein that binds to methylated H3K9 as well. The structure also reveals that the human MPP8 chromodomain forms homodimer, which is mediated via an unexpected domain swapping interaction through two β strands from the two protomer subunits. Our findings reveal the molecular mechanism of selective binding of human MPP8 chromodomain to methylated histone H3K9. The observation of human MPP8 chromodomain in both solution and crystal lattice may provide clues to study MPP8-mediated gene regulation furthermore.
Organizational Affiliation:
Key Laboratory of Structural Biology, Chinese Academy of Sciences, and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui, People's Republic of China.