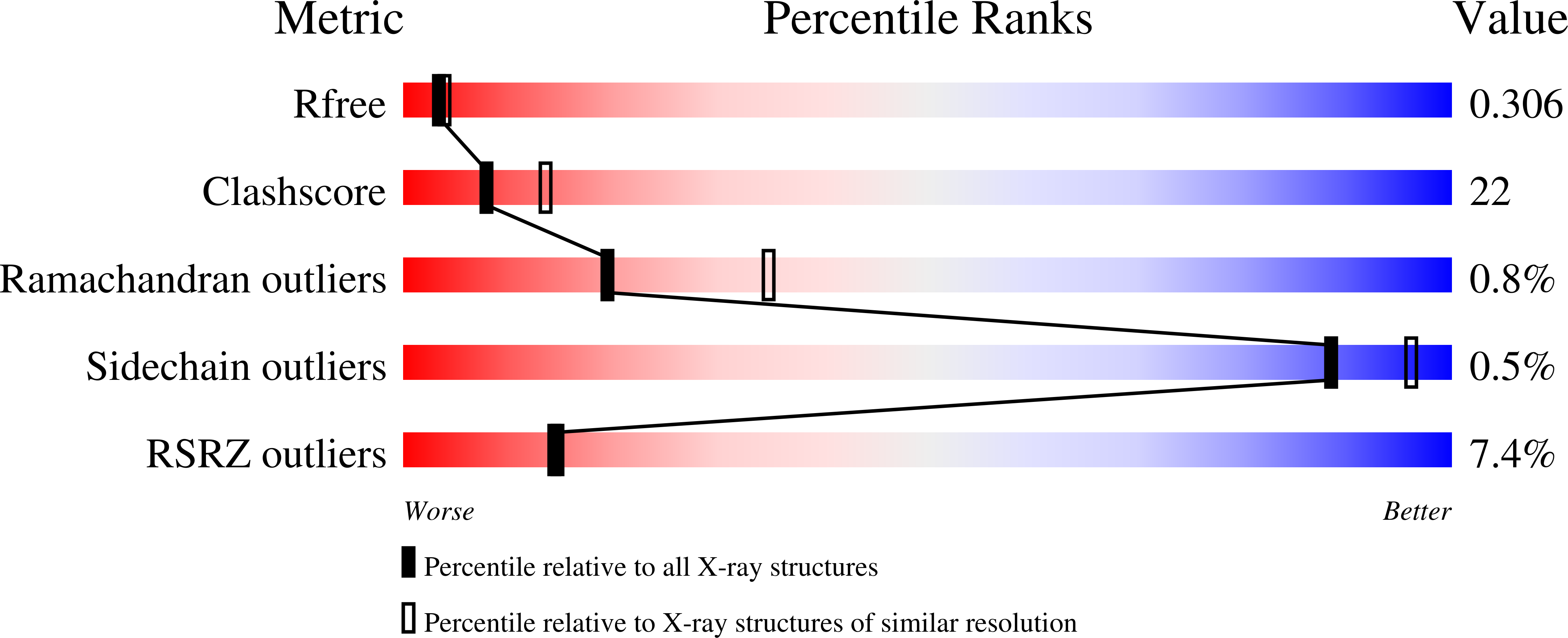
A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Kerry, P.S., Ayllon, J., Taylor, M.A., Hass, C., Lewis, A., Garcia-Sastre, A., Randall, R.E., Hale, B.G., Russell, R.J.(2011) PLoS One 6: e17946-e17946
- PubMed: 21464929
- DOI: https://doi.org/10.1371/journal.pone.0017946
- Primary Citation of Related Structures:
3O9Q, 3O9R, 3O9S, 3O9T, 3O9U, 3OA9 - PubMed Abstract:
Influenza A virus NS1 protein is a multifunctional virulence factor consisting of an RNA binding domain (RBD), a short linker, an effector domain (ED), and a C-terminal 'tail'. Although poorly understood, NS1 multimerization may autoregulate its actions. While RBD dimerization seems functionally conserved, two possible apo ED dimers have been proposed (helix-helix and strand-strand). Here, we analyze all available RBD, ED, and full-length NS1 structures, including four novel crystal structures obtained using EDs from divergent human and avian viruses, as well as two forms of a monomeric ED mutant. The data reveal the helix-helix interface as the only strictly conserved ED homodimeric contact. Furthermore, a mutant NS1 unable to form the helix-helix dimer is compromised in its ability to bind dsRNA efficiently, implying that ED multimerization influences RBD activity. Our bioinformatical work also suggests that the helix-helix interface is variable and transient, thereby allowing two ED monomers to twist relative to one another and possibly separate. In this regard, we found a mAb that recognizes NS1 via a residue completely buried within the ED helix-helix interface, and which may help highlight potential different conformational populations of NS1 (putatively termed 'helix-closed' and 'helix-open') in virus-infected cells. 'Helix-closed' conformations appear to enhance dsRNA binding, and 'helix-open' conformations allow otherwise inaccessible interactions with host factors. Our data support a new model of NS1 regulation in which the RBD remains dimeric throughout infection, while the ED switches between several quaternary states in order to expand its functional space. Such a concept may be applicable to other small multifunctional proteins.
Organizational Affiliation:
Biomedical Sciences Research Complex, University of St Andrews, St Andrews, Fife, United Kingdom.