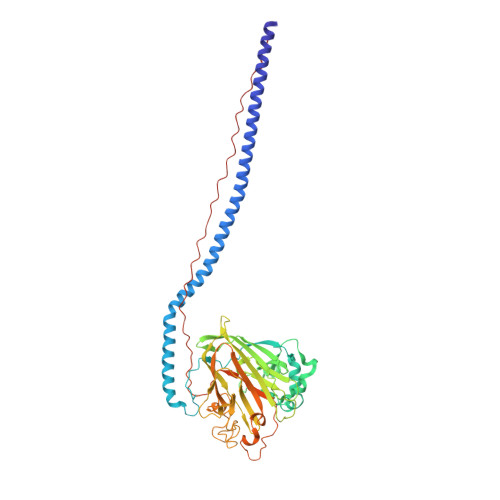
Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices.
Larson, M.R., Rajashankar, K.R., Patel, M.H., Robinette, R.A., Crowley, P.J., Michalek, S., Brady, L.J., Deivanayagam, C.(2010) Proc Natl Acad Sci U S A 107: 5983-5988
- PubMed: 20231452
- DOI: https://doi.org/10.1073/pnas.0912293107
- Primary Citation of Related Structures:
3IOX, 3IPK - PubMed Abstract:
Streptococcus mutans antigen I/II (AgI/II) is a cell surface-localized protein adhesin that interacts with salivary components within the salivary pellicle. AgI/II contributes to virulence and has been studied as an immunological and structural target, but a fundamental understanding of its underlying architecture has been lacking. Here we report a high-resolution (1.8 A) crystal structure of the A(3)VP(1) fragment of S. mutans AgI/II that demonstrates a unique fibrillar form (155 A) through the interaction of two noncontiguous regions in the primary sequence. The A(3) repeat of the alanine-rich domain adopts an extended alpha-helix that intertwines with the P(1) repeat polyproline type II (PPII) helix to form a highly extended stalk-like structure heretofore unseen in prokaryotic or eukaryotic protein structures. Velocity sedimentation studies indicate that full-length AgI/II that contains three A/P repeats extends over 50 nanometers in length. Isothermal titration calorimetry revealed that the high-affinity association between the A(3) and P(1) helices is enthalpically driven. Two distinct binding sites on AgI/II to the host receptor salivary agglutinin (SAG) were identified by surface plasmon resonance (SPR). The current crystal structure reveals that AgI/II family proteins are extended fibrillar structures with the number of alanine- and proline-rich repeats determining their length.
Organizational Affiliation:
Department of Physiology and Biophysics, University of Alabama, Birmingham, AL 35294, USA.