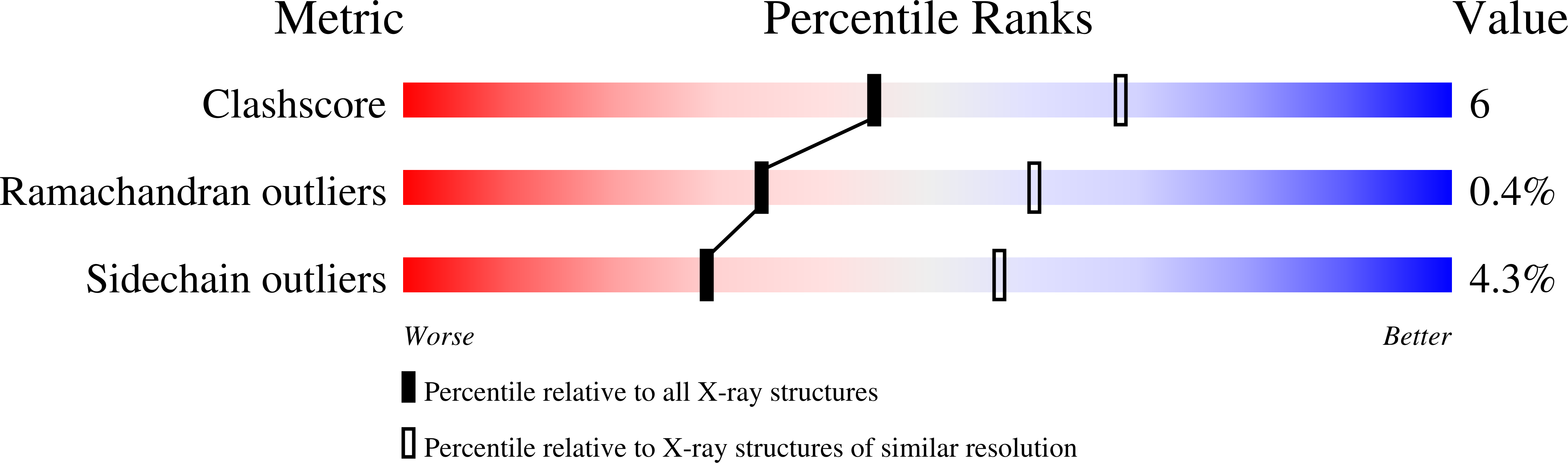
Role of an invariant lysine residue in folate binding on Escherichia coli thymidylate synthase: Calorimetric and crystallographic analysis of the K48Q mutant.
Arvizu-Flores, A.A., Sugich-Miranda, R., Arreola, R., Garcia-Orozco, K.D., Velazquez-Contreras, E.F., Montfort, W.R., Maley, F., Sotelo-Mundo, R.R.(2008) Int J Biochem Cell Biol 40: 2206-2217
- PubMed: 18403248
- DOI: https://doi.org/10.1016/j.biocel.2008.02.025
- Primary Citation of Related Structures:
2VET, 2VF0, 3B5B - PubMed Abstract:
Thymidylate synthase (TS) catalyzes the reductive methylation of deoxyuridine monophosphate (dUMP) using methylene tetrahydrofolate (CH(2)THF) as cofactor, the glutamate tail of which forms a water-mediated hydrogen bond with an invariant lysine residue of this enzyme. To understand the role of this interaction, we studied the K48Q mutant of Escherichia coli TS using structural and biophysical methods. The k(cat) of the K48Q mutant was 430-fold lower than wild-type TS in activity, while the K(m) for the (R)-stereoisomer of CH(2)THF was 300 microM, about 30-fold larger than K(m) from the wild-type TS. Affinity constants were determined using isothermal titration calorimetry, which showed that binding was reduced by one order of magnitude for folate-like TS inhibitors, such as propargyl-dideazafolate (PDDF) or compounds that distort the TS active site like BW1843U89 (U89). The crystal structure of the K48Q-dUMP complex revealed that dUMP binding is not impaired in the mutant, and that U89 in a ternary complex of K48Q-nucleotide-U89 was bound in the active site with subtle differences relative to comparable wild-type complexes. PDDF failed to form ternary complexes with K48Q and dUMP. Thermodynamic data correlated with the structural determinations, since PDDF binding was dominated by enthalpic effects while U89 had an important entropic component. In conclusion, K48 is critical for catalysis since it leads to a productive CH(2)THF binding, while mutation at this residue does not affect much the binding of inhibitors that do not make contact with this group.
Organizational Affiliation:
Aquatic Molecular Biology Laboratory, Centro de Investigación en Alimentación y Desarrollo, A.C. Hermosillo, Sonora 83000, Mexico.