Structural Analysis of the L-Alanoyl-D-Glutamate Endopeptidase Domain of Listeria Bacteriophage Endolysin Ply500 Reveals a New Member of the Las Peptidase Family.
Korndorfer, I.P., Kanitz, A., Danzer, J., Zimmer, M., Loessner, M.J., Skerra, A.(2008) Acta Crystallogr D Biol Crystallogr 64: 644
- PubMed: 18560152
- DOI: https://doi.org/10.1107/S0907444908007890
- Primary Citation of Related Structures:
2VO9 - PubMed Abstract:
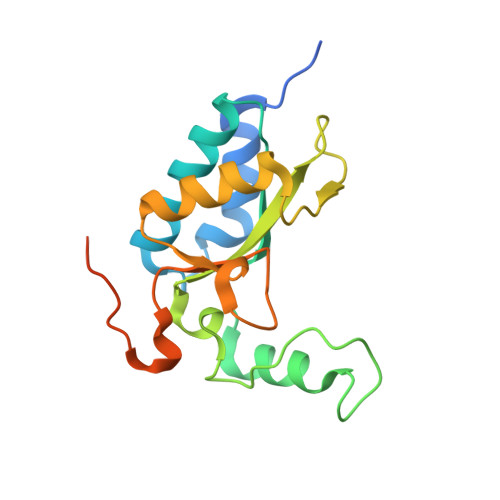
Similar to many other bacterial cell-wall-hydrolyzing enzymes, the Listeria bacteriophage A500 endopeptidase Ply500 has a modular architecture consisting of an enzymatically active domain (EAD) linked to a cell-wall-binding domain (CBD) in a single polypeptide chain. The crystal structure of the EAD of Ply500 has been solved at 1.8 A resolution. The shape of the enzyme resembles a sofa chair: one alpha-helix and three antiparallel beta-strands form the seat, which is supported by two more alpha-helices, while another alpha-helix together with the following loop give rise to the backrest. A sulfate anion bound to the active site, which harbours a catalytic Zn2+ ion, indicates mechanistic details of the peptidase reaction, which involves a tetrahedral transition state. Despite very low sequence similarity, a clear structural relationship was detected to the peptidases VanX, DDC, MSH and MepA, which belong to the so-called 'LAS' family. Their gross functional similarity is supported by a common bound Zn2+ ion and a highly conserved set of coordinating residues (His80, Asp87 and His133) as well as other side chains (Arg50, Gln55, Ser78 and Asp130) in the active site. Considering the high sequence similarity to the EAD of the Listeria phage endopeptidase Ply118, both enzymes can thus be assigned to the LAS family. The same is the case for the L,D-endopeptidase CwlK from Bacillus subtilis, which shows both functional and amino-acid sequence similarity. The fact that the CBD of Ply500 is closely homologous to the CBD of the Listeria phage N-acetylmuramoyl-L-alanine amidase PlyPSA, which exhibits a totally different EAD, illustrates the modular composition and functional variability of this class of enzymes and opens interesting possibilities for protein engineering.
Organizational Affiliation:
Lehrstuhl für Biologische Chemie, Technische Universität München, An der Saatzucht 5, D-85350 Freising, Germany. korndoerfer@crelux.com