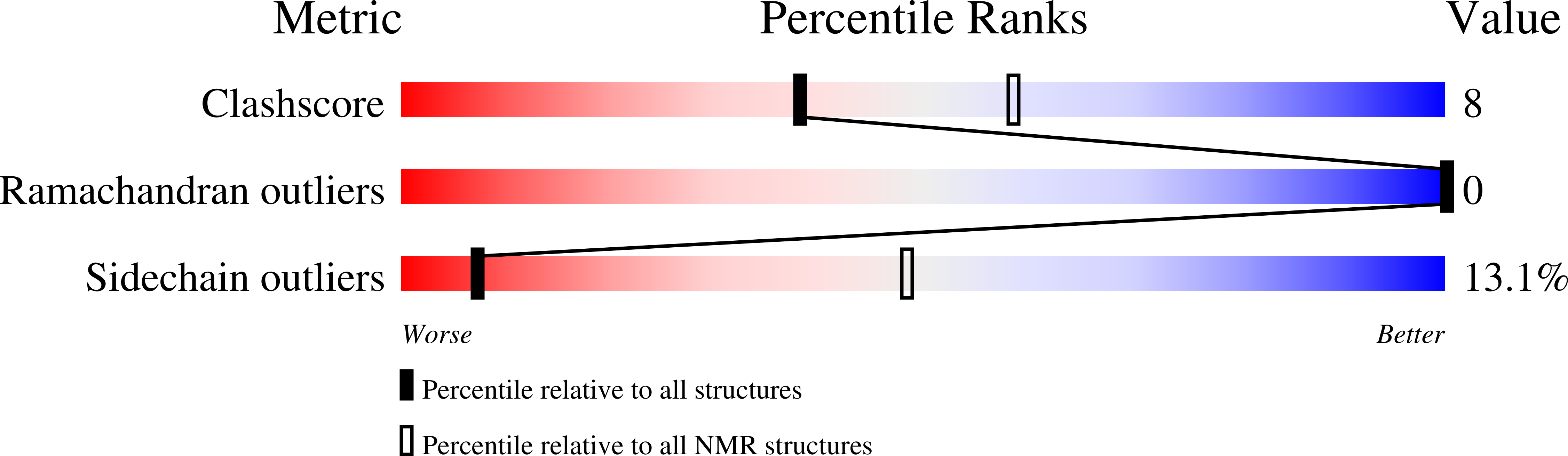Structure/Function Analysis of Protein-Protein Interactions Developed by the Yeast Pih1 Platform Protein and Its Partners in Box C/D snoRNP Assembly.
Quinternet, M., Rothe, B., Barbier, M., Bobo, C., Saliou, J.M., Jacquemin, C., Back, R., Chagot, M.E., Cianferani, S., Meyer, P., Branlant, C., Charpentier, B., Manival, X.(2015) J Mol Biol 427: 2816-2839
- PubMed: 26210662
- DOI: https://doi.org/10.1016/j.jmb.2015.07.012
- Primary Citation of Related Structures:
2MNJ - PubMed Abstract:
In eukaryotes, nucleotide post-transcriptional modifications in RNAs play an essential role in cell proliferation by contributing to pre-ribosomal RNA processing, ribosome assembly and activity. Box C/D small nucleolar ribonucleoparticles catalyze site-specific 2'-O-methylation of riboses, one of the most prevalent RNA modifications. They contain one guide RNA and four core proteins and their in vivo assembly requires numerous factors including (HUMAN/Yeast) BCD1/Bcd1p, NUFIP1/Rsa1p, ZNHIT3/Hit1p, the R2TP complex composed of protein PIH1D1/Pih1p and RPAP3/Tah1p that bridges the R2TP complex to the HSP90/Hsp82 chaperone and two AAA+ ATPases. We show that Tah1p can stabilize Pih1p in the absence of Hsp82 activity during the stationary phase of growth and consequently that the Tah1p:Pih1p interaction is sufficient for Pih1p stability. This prompted us to establish the solution structure of the Tah1p:Pih1p complex by NMR. The C-terminal tail S93-S111 of Tah1p snakes along Pih1p264-344 folded in a CS domain to form two intermolecular β-sheets and one covering loop. However, a thorough inspection of the NMR and crystal structures revealed structural differences that may be of functional importance. In addition, our NMR and isothermal titration calorimetry data revealed the formation of direct contacts between Pih1p257-344 and the Hsp82MC domain in the presence of Tah1p. By co-expression in Escherichia coli, we demonstrate that Pih1p has two other direct partners, the Rsa1p assembly factor and the Nop58p core protein, and in vivo and in vitro experiments mapped the required binding domains. Our data suggest that these two interactions are mutually exclusive. The implication of this finding for box C/D small nucleolar ribonucleoparticle assembly is discussed.
Organizational Affiliation:
FR 3209 CNRS-Université de Lorraine, Bioingénierie Moléculaire, Cellulaire et Thérapeutique, Biopôle, Campus Biologie-Santé, CS 50184, 54505 Vandœuvre-lès-Nancy Cedex, France.