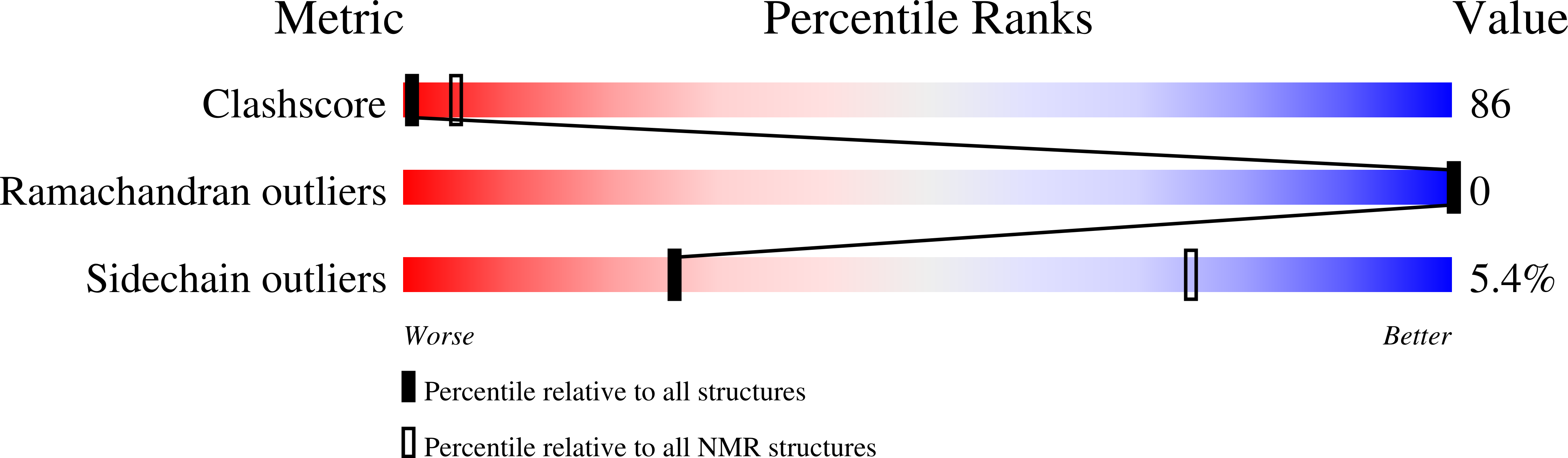Structural and functional characterization of alternative transmembrane domain conformations in VEGF receptor 2 activation
Manni, S., Mineev, K.S., Usmanova, D., Lyukmanova, E.N., Shulepko, M.A., Kirpichnikov, M.P., Winter, J., Matkovic, M., Deupi, X., Arseniev, A.S., Ballmer-Hofer, K.(2014) Structure 22: 1077-1089
- PubMed: 24980797
- DOI: https://doi.org/10.1016/j.str.2014.05.010
- Primary Citation of Related Structures:
2M59, 2MET, 2MEU - PubMed Abstract:
Transmembrane signaling by receptor tyrosine kinases (RTKs) entails ligand-mediated dimerization and structural rearrangement of the extracellular domains. RTK activation also depends on the specific orientation of the transmembrane domain (TMD) helices, as suggested by pathogenic, constitutively active RTK mutants. Such mutant TMDs carry polar amino acids promoting stable transmembrane helix dimerization, which is essential for kinase activation. We investigated the effect of polar amino acids introduced into the TMD of vascular endothelial growth factor receptor 2, regulating blood vessel homeostasis. Two mutants showed constitutive kinase activity, suggesting that precise TMD orientation is mandatory for kinase activation. Nuclear magnetic resonance spectroscopy revealed that TMD helices in activated constructs were rotated by 180° relative to the interface of the wild-type conformation, confirming that ligand-mediated receptor activation indeed results from transmembrane helix rearrangement. A molecular dynamics simulation confirmed the transmembrane helix arrangement of wild-type and mutant TMDs revealed by nuclear magnetic resonance spectroscopy.
Organizational Affiliation:
Paul Scherrer Institute, Biomolecular Research, 5232 Villigen PSI, Switzerland.