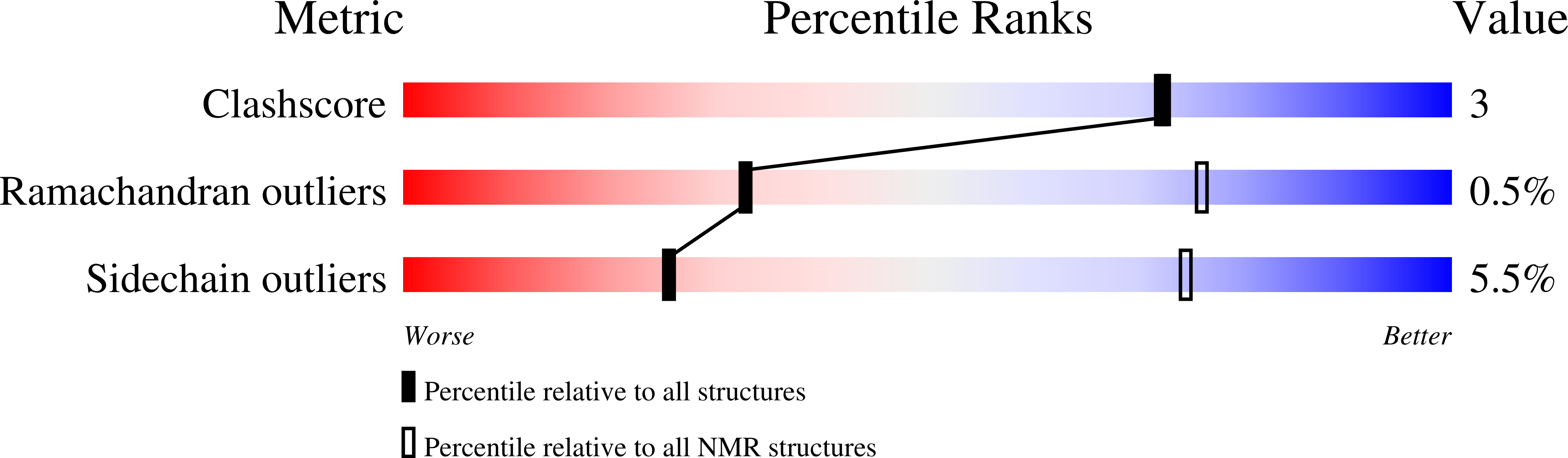Retinoblastoma-binding protein 1 has an interdigitated double Tudor domain with DNA binding activity.
Gong, W., Wang, J., Perrett, S., Feng, Y.(2014) J Biol Chem 289: 4882-4895
- PubMed: 24379399
- DOI: https://doi.org/10.1074/jbc.M113.501940
- Primary Citation of Related Structures:
2MAM - PubMed Abstract:
Retinoblastoma-binding protein 1 (RBBP1) is a tumor and leukemia suppressor that binds both methylated histone tails and DNA. Our previous studies indicated that RBBP1 possesses a Tudor domain, which cannot bind histone marks. In order to clarify the function of the Tudor domain, the solution structure of the RBBP1 Tudor domain was determined by NMR and is presented here. Although the proteins are unrelated, the RBBP1 Tudor domain forms an interdigitated double Tudor structure similar to the Tudor domain of JMJD2A, which is an epigenetic mark reader. This indicates the functional diversity of Tudor domains. The RBBP1 Tudor domain structure has a significant area of positively charged surface, which reveals a capability of the RBBP1 Tudor domain to bind nucleic acids. NMR titration and isothermal titration calorimetry experiments indicate that the RBBP1 Tudor domain binds both double- and single-stranded DNA with an affinity of 10-100 μM; no apparent DNA sequence specificity was detected. The DNA binding mode and key interaction residues were analyzed in detail based on a model structure of the Tudor domain-dsDNA complex, built by HADDOCK docking using the NMR data. Electrostatic interactions mediate the binding of the Tudor domain with DNA, which is consistent with NMR experiments performed at high salt concentration. The DNA-binding residues are conserved in Tudor domains of the RBBP1 protein family, resulting in conservation of the DNA-binding function in the RBBP1 Tudor domains. Our results provide further insights into the structure and function of RBBP1.
Organizational Affiliation:
From the National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China and.