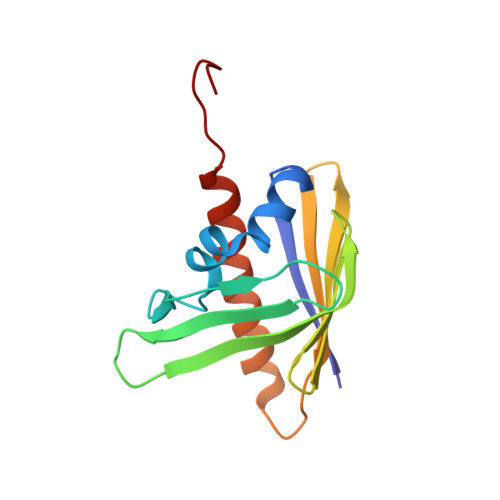
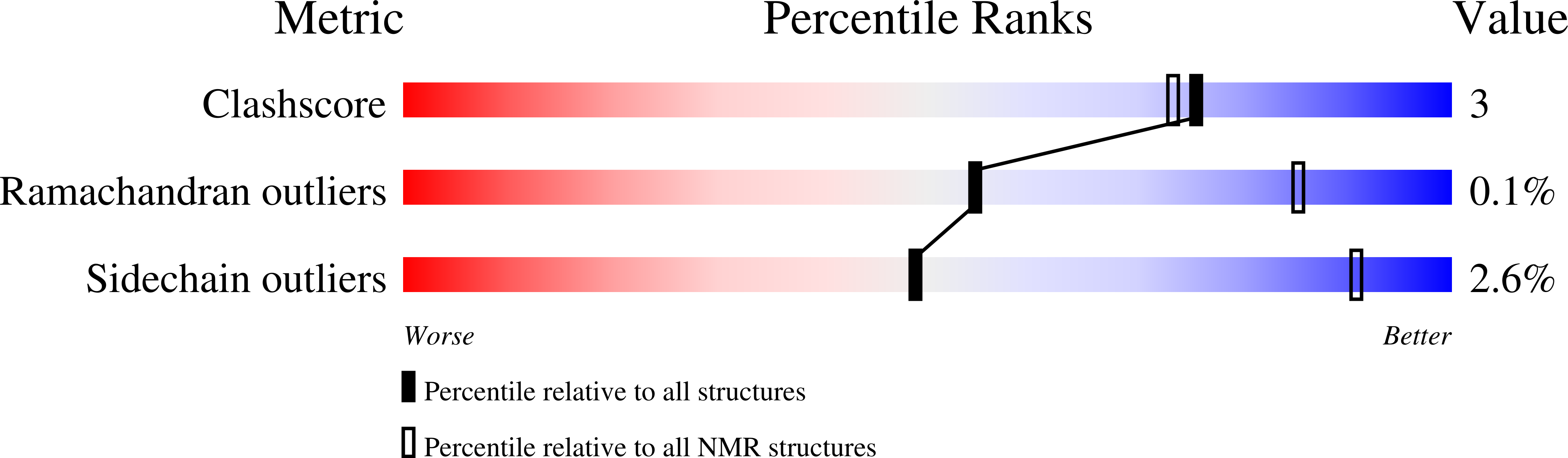Solution NMR structure of protein Rmet_5065 from Ralstonia metallidurans. Northeast Structural Genomics Consortium Target CrR115.
Ramelot, T.A., Yang, Y., Cort, J.R., Wang, D., Ciccosanti, C., Janjua, H., Rajesh, N., Acton, T.B., Xiao, R., Everett, J.K., Montelione, G.T., Kennedy, M.A.To be published.