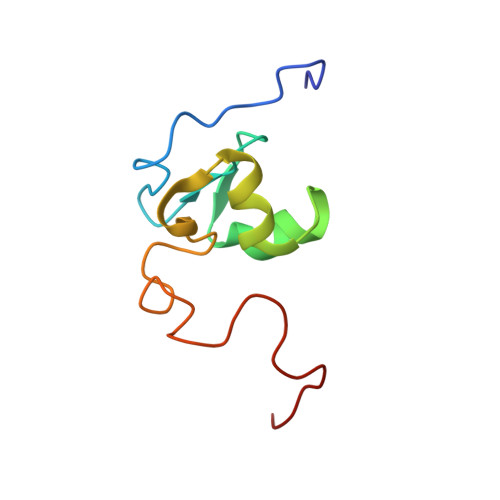
The prokaryotic Cys2His2 zinc-finger adopts a novel fold as revealed by the NMR structure of Agrobacterium tumefaciens Ros DNA-binding domain.
Malgieri, G., Russo, L., Esposito, S., Baglivo, I., Zaccaro, L., Pedone, E.M., Di Blasio, B., Isernia, C., Pedone, P.V., Fattorusso, R.(2007) Proc Natl Acad Sci U S A 104: 17341-17346
- PubMed: 17956987
- DOI: https://doi.org/10.1073/pnas.0706659104
- Primary Citation of Related Structures:
2JSP - PubMed Abstract:
The first putative prokaryotic Cys(2)His(2) zinc-finger domain has been identified in the transcriptional regulator Ros from Agrobacterium tumefaciens, indicating that the Cys(2)His(2) zinc-finger domain, originally thought to be confined to the eukaryotic kingdom, could be widespread throughout the living kingdom from eukaryotic, both animal and plant, to prokaryotic. In this article we report the NMR solution structure of Ros DNA-binding domain (Ros87), providing 79 structural characterization of a prokaryotic Cys(2)His(2) zinc-finger domain. The NMR structure of Ros87 shows that the putative prokaryotic Cys(2)His(2) zinc-finger sequence is indeed part of a significantly larger zinc-binding globular domain that possesses a novel protein fold very different from the classical fold reported for the eukaryotic classical zinc-finger. The Ros87 globular domain consists of 58 aa (residues 9-66), is arranged in a betabetabetaalphaalpha topology, and is stabilized by an extensive 15-residue hydrophobic core. A backbone dynamics study of Ros87, based on (15)N R(1), (15)N R(2), and heteronuclear (15)N-{(1)H}-NOE measurements, has further confirmed that the globular domain is uniformly rigid and flanked by two flexible tails. Mapping of the amino acids necessary for the DNA binding onto Ros87 structure reveals the protein surface involved in the DNA recognition mechanism of this new zinc-binding protein domain.
Organizational Affiliation:
Dipartimento di Scienze Ambientali, Seconda Università degli Studi di Napoli, Via Vivaldi 43, 81100 Caserta, Italy.