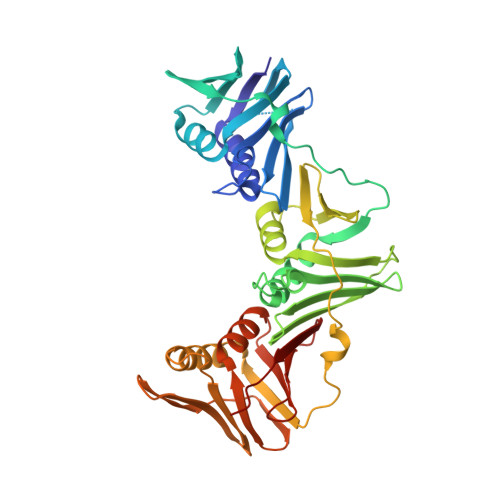
Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium.
Argiriadi, M.A., Goedken, E.R., Bruck, I., O'donnell, M., Kuriyan, J.(2006) BMC Struct Biol 6: 2-2
- PubMed: 16403212
- DOI: https://doi.org/10.1186/1472-6807-6-2
- Primary Citation of Related Structures:
2AVT - PubMed Abstract:
Sliding DNA clamps are processivity factors that are required for efficient DNA replication. DNA polymerases maintain proximity to nucleic acid templates by interacting with sliding clamps that encircle DNA and thereby link the polymerase enzyme to the DNA substrate. Although the structures of sliding clamps from Gram-negative bacteria (E. coli), eukaryotes, archaea, and T4-like bacteriophages are well-known, the structure of a sliding clamp from Gram-positive bacteria has not been reported previously. We have determined the crystal structure of the dimeric beta subunit of the DNA polymerase III holoenzyme of Streptococcus pyogenes. The sliding clamp from this Gram-positive organism forms a ring-shaped dimeric assembly that is similar in overall structure to that of the sliding clamps from Gram-negative bacteria, bacteriophage T4, eukaryotes and archaea. The dimer has overall dimensions of approximately 90 A x approximately 70 A x approximately 25 A with a central chamber that is large enough to accommodate duplex DNA. In comparison to the circular shape of other assemblies, the S. pyogenes clamp adopts a more elliptical structure. The sequences of sliding clamps from S. pyogenes and E. coli are only 23% identical, making the generation of structural models for the S. pyogenes clamp difficult in the absence of direct experimental information. Our structure of the S. pyogenes beta subunit completes the catalog of clamp structures from all the major sequence grouping of sliding clamps. The more elliptical rather than circular structure of the S. pyogenes clamp implies that the topological nature of encircling DNA, rather than a precise geometric shape, is the most conserved aspect for this family of proteins.
Organizational Affiliation:
Howard Hughes Medical Institute, The Rockefeller University, New York, NY 10021, USA. maria.argiriadi@abbott.com