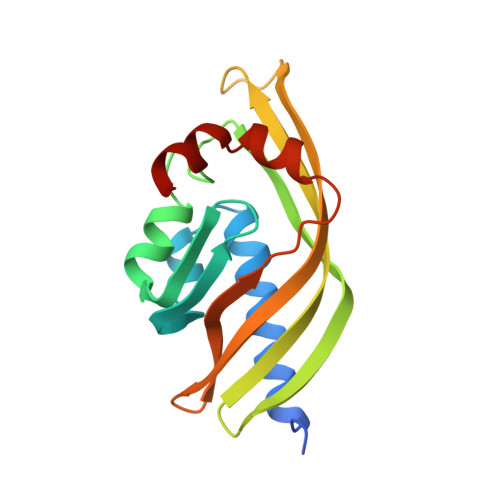
Crystal structure of scytalone dehydratase--a disease determinant of the rice pathogen, Magnaporthe grisea.
Lundqvist, T., Rice, J., Hodge, C.N., Basarab, G.S., Pierce, J., Lindqvist, Y.(1994) Structure 2: 937-944
- PubMed: 7866745
- DOI: https://doi.org/10.1016/s0969-2126(94)00095-6
- Primary Citation of Related Structures:
1STD - PubMed Abstract:
Rice blast is caused by the pathogenic fungus,-Magnaporthe grisea. Non-pathogenic mutants have been identified that lack enzymes in the biosynthetic pathway of dihydroxynapthalene-derived melanin. These enzymes are therefore prime targets for fungicides designed to control rice blast disease. One of the enzymes identified by genetic analysis as a disease determinant is scytalone dehydratase. The three-dimensional structure of scytalone dehydratase in complex with a competitive inhibitor has been determined at 2.9 A resolution. A novel fold, a cone-shaped alpha + beta barrel, is adopted by the monomer in this trimeric protein, burying the hydrophobic active site in its interior. The interactions of the inhibitor with the protein side chains have been identified. The similarity of the inhibitor to the substrate and the side chains involved in binding afford some insights into possible catalytic mechanisms. These results provide a first look into the structure and catalytic residues of a non-metal dehydratase, a large class of hitherto structurally uncharacterized enzymes. It is envisaged that a detailed structural description of scytalone dehydratase will assist in the design of new inhibitors for controlling rice blast disease.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences Uppsala Biomedical Center.