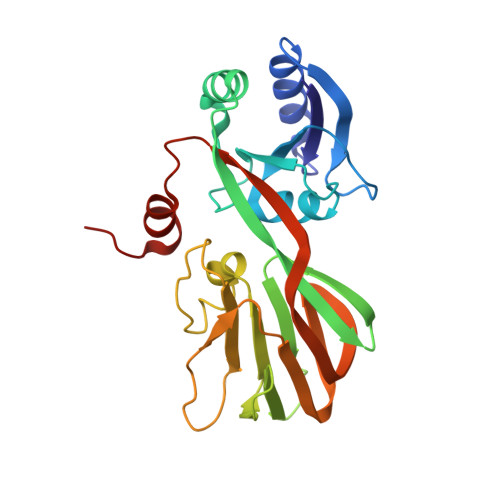
Structure of a NAD kinase from Thermotoga maritima at 2.3 A resolution.
Oganesyan, V., Huang, C., Adams, P.D., Jancarik, J., Yokota, H.A., Kim, R., Kim, S.H.(2005) Acta Crystallogr Sect F Struct Biol Cryst Commun 61: 640-646
- PubMed: 16511117
- DOI: https://doi.org/10.1107/S1744309105019780
- Primary Citation of Related Structures:
1YT5 - PubMed Abstract:
NAD kinase is the only known enzyme that catalyzes the formation of NADP, a coenzyme involved in most anabolic reactions and in the antioxidant defense system. Despite its importance, very little is known regarding the mechanism of catalysis and only recently have several NAD kinase structures been deposited in the PDB. Here, an independent investigation of the crystal structure of inorganic polyphosphate/ATP-NAD kinase, PPNK_THEMA, a protein from Thermotoga maritima, is reported at a resolution of 2.3 A. The crystal structure was solved using single-wavelength anomalous diffraction (SAD) data collected at the Se absorption-peak wavelength in a state in which no cofactors or substrates were bound. It revealed that the 258-amino-acid protein is folded into two distinct domains, similar to recently reported NAD kinases. The N-terminal alpha/beta-domain spans the first 100 amino acids and the last 30 amino acids of the polypeptide and has several topological matches in the PDB, whereas the other domain, which spans the middle 130 residues, adopts a unique beta-sandwich architecture and only appreciably matches the recently deposited PDB structures of NAD kinases.
Organizational Affiliation:
Structural Genomics Center, Physical Biosciences Division, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA.