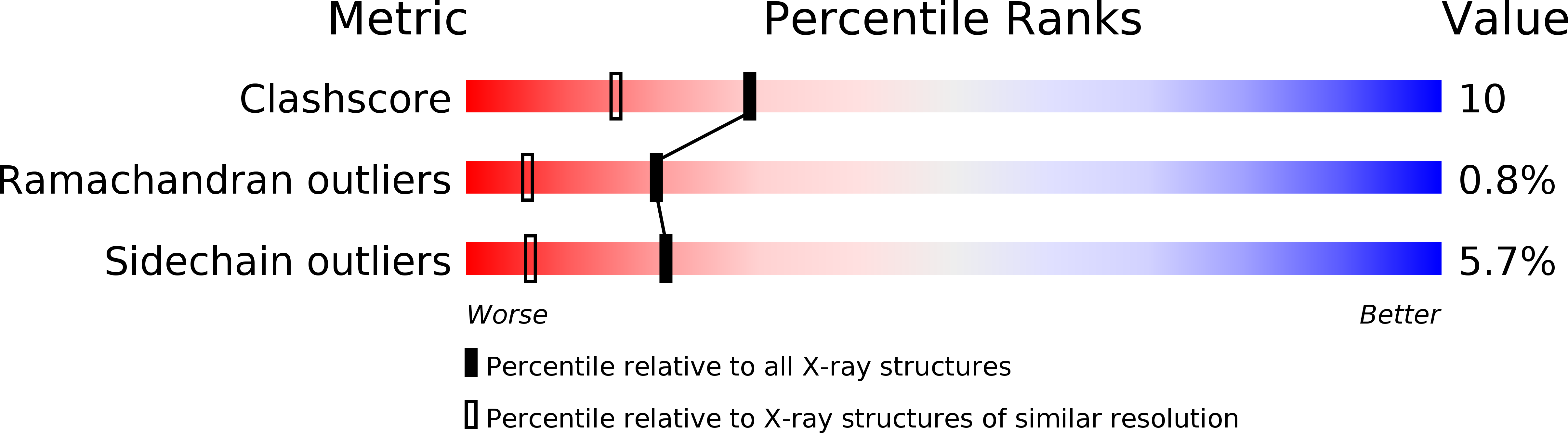Identification and structural analysis of a zebrafish apo and holo cellular retinol-binding protein.
Calderone, V., Folli, C., Marchesani, A., Berni, R., Zanotti, G.(2002) J Mol Biol 321: 527-535
- PubMed: 12162964
- DOI: https://doi.org/10.1016/s0022-2836(02)00628-9
- Primary Citation of Related Structures:
1KQW, 1KQX - PubMed Abstract:
Cellular retinol-binding proteins (CRBPs) are cytoplasmic retinol-specific binding proteins. Mammalian CRBPs have been thoroughly characterised previously. Here we report on the identification and X-ray structural analysis of the apo (1.7A resolution) and holo (1.4A resolution) forms of a zebrafish CRBP. According to amino acid sequence and structure analyses, the zebrafish CRBP that we have identified resembles closely mammalian CRBP II, suggesting that it is the zebrafish orthologue of this mammalian CRBP type. Zebrafish CRBP forms a tight complex with all-trans retinol, producing an absorption spectrum similar to those of mammalian holo-CRBPs, albeit slightly blue-shifted. The superposition of the alpha-carbon atoms of the liganded (complexed with retinol) and unliganded forms of zebrafish CRBP shows significant differences in correspondence of the betaC-betaD (residues 55-58) and betaE-betaF (residues 74-77) turns, providing evidence for the occurrence of conformational changes accompanying retinol binding/release. Remarkable and well-defined ligand-dependent conformational changes in the protein region comprising the two beta-turns affect both the main chain and the side-chains of several residues. The two beta-turns project towards the interior of the cavity devoid of ligand of the apoprotein. The side-chains of F57, Y60 and L77 change substantially their orientation and position in the apoprotein relative to the holoprotein. In the beta-barrel internal cavity of apo-CRBP they occupy some of the space that is otherwise occupied by bound retinol in holo-CRBP, and are displaced from these positions on ligand binding. These results indicate that a flexible area encompassing the betaC-betaD and betaE-betaF turns may serve as the ligand portal and that these turns undergo conformational changes associated with the not yet clarified mechanism of retinol binding and release in CRBPs.
Organizational Affiliation:
Department of Organic Chemistry, University of Padova, and Biopolymer Research Center, Italian National Research Council, Via Marzolo 1, 35131, Padova, Italy.