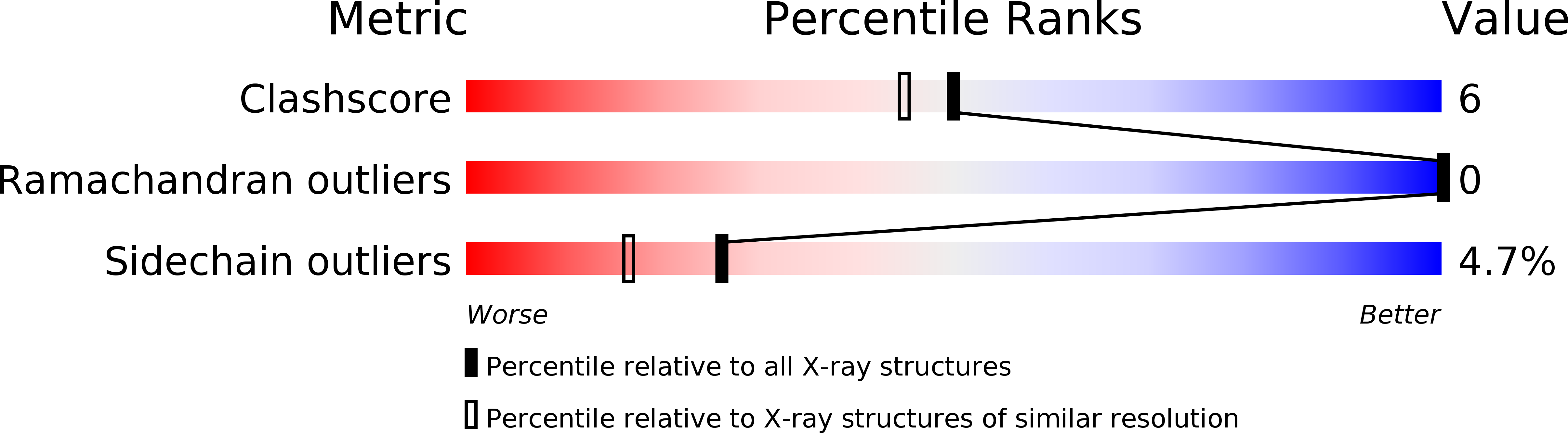Structure of a microbial homologue of mammalian platelet-activating factor acetylhydrolases: Streptomyces exfoliatus lipase at 1.9 A resolution.
Wei, Y., Swenson, L., Castro, C., Derewenda, U., Minor, W., Arai, H., Aoki, J., Inoue, K., Servin-Gonzalez, L., Derewenda, Z.S.(1998) Structure 6: 511-519
- PubMed: 9562561
- DOI: https://doi.org/10.1016/s0969-2126(98)00052-5
- Primary Citation of Related Structures:
1JFR - PubMed Abstract:
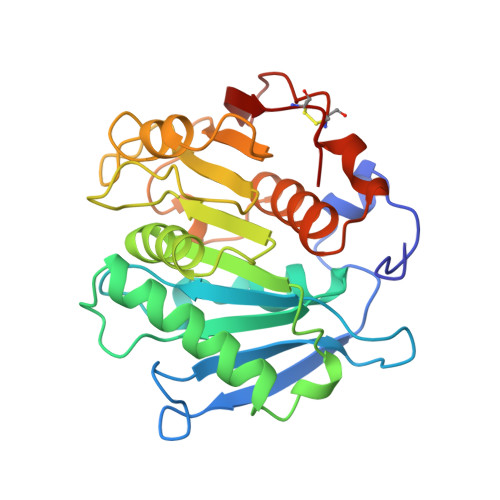
Neutral lipases are ubiquitous and diverse enzymes. The molecular architecture of the structurally characterized lipases is similar, often despite a lack of detectable homology at the sequence level. Some of the microbial lipases are evolutionarily related to physiologically important mammalian enzymes. For example, limited sequence similarities were recently noted for the Streptomyces exfoliatus lipase (SeL) and two mammalian platelet-activating factor acetylhydrolases (PAF-AHs). The determination of the crystal structure of SeL allowed us to explore the structure-function relationships in this novel family of homologous hydrolases. The crystal structure of SeL was determined by multiple isomorphous replacement and refined using data to 1.9 A resolution. The molecule exhibits the canonical tertiary fold of an alpha/beta hydrolase. The putative nucleophilic residue, Ser131, is located within a nucleophilic elbow and is hydrogen bonded to His209, which in turn interacts with Asp177. These three residues create a triad that closely resembles the catalytic triads found in the active sites of other neutral lipases. The mainchain amides of Met132 and Phe63 are perfectly positioned to create an oxyanion hole. Unexpectedly, there are no secondary structure elements that could render the active site inaccessible to solvent, like the lids that are commonly found in neutral lipases. The crystal structure of SeL reinforces the notion that it is a homologue of the mammalian PAF-AHs. We have used the catalytic triad in SeL to model the active site of the PAF-AHs. Our model is consistent with the site-directed mutagenesis studies of plasma PAF-AH, which implicate Ser273, His351 and Asp296 in the active site. Our study therefore provides direct support for the hypothesis that the plasma and isoform II PAF-AHs are triad-containing alpha/beta hydrolases.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia, Health Sciences Center, Charlottesville, VA 22906, USA.