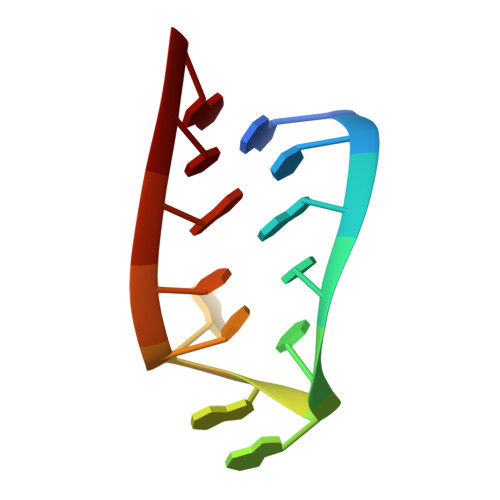
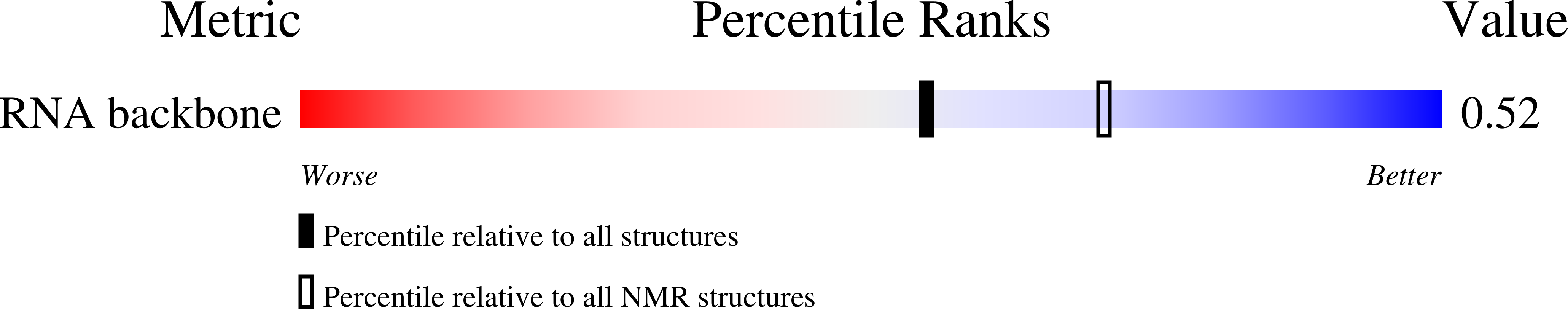Structural characterization of a six-nucleotide RNA hairpin loop found in Escherichia coli, r(UUAAGU).
Zhang, H., Fountain, M.A., Krugh, T.R.(2001) Biochemistry 40: 9879-9886
- PubMed: 11502181
- DOI: https://doi.org/10.1021/bi011226x
- Primary Citation of Related Structures:
1HS2 - PubMed Abstract:
The binding region of the Escherichia coli S2 ribosomal protein contains a conserved UUAAGU hairpin loop. The structure of the hairpin formed by the oligomer r(GCGU4U5A6A7G8U9CGCA), which has an r(UUAAGU) hairpin loop, was determined by NMR and molecular modeling techniques as part of a study aimed at characterizing the structure and thermodynamics of RNA hairpin loops. Thermodynamic data obtained from melting curves for this RNA oligomer show that it forms a hairpin in solution with the following parameters: DeltaH degrees = -42.8 +/- 2.2 kcal/mol, DeltaS degrees = -127.6 +/- 6.5 eu, and DeltaG degrees (37) = -3.3 +/- 0.2 kcal/mol. Two-dimensional NOESY WATERGATE spectra show an NOE between U imino protons, which suggests that U4 and U9 form a hydrogen bonded U.U pair. The U5(H2') proton shows NOEs to both the A6(H8) proton and the A7(H8) proton, which is consistent with formation of a "U" turn between nucleotides U5 and A6. An NOE between the A7(H2) proton and the U9(H4') proton shows the proximity of the A7 base to the U9 sugar, which is consistent with the structure determined for the six-nucleotide loop. In addition to having a hydrogen-bonded U.U pair as the first mismatch and a U turn, the r(UUAAGU) loop has the G8 base protruding into the solvent. The solution structure of the r(UUAAGU) loop is essentially identical to the structure of an identical loop found in the crystal structure of the 30S ribosomal subunit where the guanine in the loop is involved in tertiary interactions with RNA bases from adjacent regions [Wimberly, B. T., Brodersen, D. E., Clemons, W. M., Morgan-Warren, R. J., Carter, A. P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. (2000) Nature 407, 327-339]. The similarity of the solution and solid-state structures of this hairpin loop suggests that formation of this hairpin may facilitate folding of 16S RNA.
Organizational Affiliation:
Department of Chemistry, University of Rochester, Rochester, New York 14627-0216, USA.